Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability
- PMID: 37821446
- PMCID: PMC10567765
- DOI: 10.1038/s41541-023-00751-6
Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability
Abstract
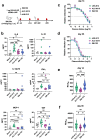
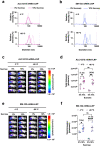
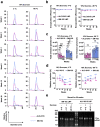
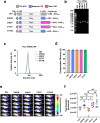
During the COVID-19 pandemic, Pfizer-BioNTech and Moderna successfully developed nucleoside-modified mRNA lipid nanoparticle (LNP) vaccines. SARS-CoV-2 spike protein expressed by those vaccines are identical in amino acid sequence, but several key components are distinct. Here, we compared the effect of ionizable lipids, untranslated regions (UTRs), and nucleotide composition of the two vaccines, focusing on mRNA delivery, antibody generation, and long-term stability. We found that the ionizable lipid, SM-102, in Moderna's vaccine performs better than ALC-0315 in Pfizer-BioNTech's vaccine for intramuscular delivery of mRNA and antibody production in mice and long-term stability at 4 °C. Moreover, Pfizer-BioNTech's 5' UTR and Moderna's 3' UTR outperform their counterparts in their contribution to transgene expression in mice. We further found that varying N1-methylpseudouridine content at the wobble position of mRNA has little effect on vaccine efficacy. These findings may contribute to the further improvement of nucleoside-modified mRNA-LNP vaccines and therapeutics.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Vogel AB, et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

