Inherently interpretable position-aware convolutional motif kernel networks for biological sequencing data
- PMID: 37821530
- PMCID: PMC10567796
- DOI: 10.1038/s41598-023-44175-7
Inherently interpretable position-aware convolutional motif kernel networks for biological sequencing data
Abstract
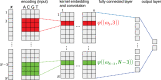
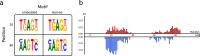
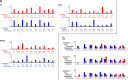
Artificial neural networks show promising performance in detecting correlations within data that are associated with specific outcomes. However, the black-box nature of such models can hinder the knowledge advancement in research fields by obscuring the decision process and preventing scientist to fully conceptualize predicted outcomes. Furthermore, domain experts like healthcare providers need explainable predictions to assess whether a predicted outcome can be trusted in high stakes scenarios and to help them integrating a model into their own routine. Therefore, interpretable models play a crucial role for the incorporation of machine learning into high stakes scenarios like healthcare. In this paper we introduce Convolutional Motif Kernel Networks, a neural network architecture that involves learning a feature representation within a subspace of the reproducing kernel Hilbert space of the position-aware motif kernel function. The resulting model enables to directly interpret and evaluate prediction outcomes by providing a biologically and medically meaningful explanation without the need for additional post-hoc analysis. We show that our model is able to robustly learn on small datasets and reaches state-of-the-art performance on relevant healthcare prediction tasks. Our proposed method can be utilized on DNA and protein sequences. Furthermore, we show that the proposed method learns biologically meaningful concepts directly from data using an end-to-end learning scheme.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

