Ultra-fast deep-learned CNS tumour classification during surgery
- PMID: 37821699
- PMCID: PMC10600004
- DOI: 10.1038/s41586-023-06615-2
Ultra-fast deep-learned CNS tumour classification during surgery
Abstract
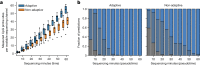
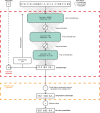
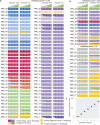
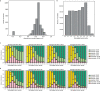
Central nervous system tumours represent one of the most lethal cancer types, particularly among children1. Primary treatment includes neurosurgical resection of the tumour, in which a delicate balance must be struck between maximizing the extent of resection and minimizing risk of neurological damage and comorbidity2,3. However, surgeons have limited knowledge of the precise tumour type prior to surgery. Current standard practice relies on preoperative imaging and intraoperative histological analysis, but these are not always conclusive and occasionally wrong. Using rapid nanopore sequencing, a sparse methylation profile can be obtained during surgery4. Here we developed Sturgeon, a patient-agnostic transfer-learned neural network, to enable molecular subclassification of central nervous system tumours based on such sparse profiles. Sturgeon delivered an accurate diagnosis within 40 minutes after starting sequencing in 45 out of 50 retrospectively sequenced samples (abstaining from diagnosis of the other 5 samples). Furthermore, we demonstrated its applicability in real time during 25 surgeries, achieving a diagnostic turnaround time of less than 90 min. Of these, 18 (72%) diagnoses were correct and 7 did not reach the required confidence threshold. We conclude that machine-learned diagnosis based on low-cost intraoperative sequencing can assist neurosurgical decision-making, potentially preventing neurological comorbidity and avoiding additional surgeries.
© 2023. The Author(s).
Conflict of interest statement
J.d.R., M.P.-G. and CV are inventors on a patent covering the development of Sturgeon. J.d.R. is co-founder and director of Cyclomics, a genomics company. L.K., M.E.G.K., P.W., N.V., P.d.W.H., E.J.K., L.D., J.v.d.L., K.v.B., E.W.H. and B.B.J.T. declare no competing interests.
Figures








Comment in
-
AI rapidly diagnoses brain tumours during surgery.Nature. 2023 Oct;622(7984):702-703. doi: 10.1038/d41586-023-03072-9. Nature. 2023. PMID: 37848711 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

