Expanding the Range of Bioorthogonal Tags for Multiplex Stimulated Raman Scattering Microscopy
- PMID: 37821742
- PMCID: PMC10952743
- DOI: 10.1002/anie.202311530
Expanding the Range of Bioorthogonal Tags for Multiplex Stimulated Raman Scattering Microscopy
Abstract
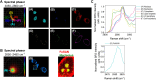
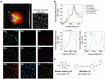
Multiplex optical detection in live cells is challenging due to overlapping signals and poor signal-to-noise associated with some chemical reporters. To address this, the application of spectral phasor analysis to stimulated Raman scattering (SRS) microscopy for unmixing three bioorthogonal Raman probes within cells is reported. Triplex detection of a metallacarborane using the B-H stretch at 2480-2650 cm-1 , together with a bis-alkyne and deuterated fatty acid can be achieved within the cell-silent region of the Raman spectrum. When coupled to imaging in the high-wavenumber region of the cellular Raman spectrum, nine discrete regions of interest can be spectrally unmixed from the hyperspectral SRS dataset, demonstrating a new capability in the toolkit of multiplexed Raman imaging of live cells.
Keywords: Bioorthogonal Labeling; Imaging Probes; Raman Microscopy; Spectral Phasor Analysis; Stimulated Raman Scattering.
© 2023 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

