The evolution of antimicrobial peptides in Chiroptera
- PMID: 37822944
- PMCID: PMC10562630
- DOI: 10.3389/fimmu.2023.1250229
The evolution of antimicrobial peptides in Chiroptera
Abstract
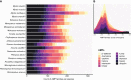
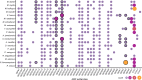
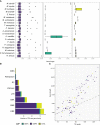
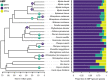
High viral tolerance coupled with an extraordinary regulation of the immune response makes bats a great model to study host-pathogen evolution. Although many immune-related gene gains and losses have been previously reported in bats, important gene families such as antimicrobial peptides (AMPs) remain understudied. We built an exhaustive bioinformatic pipeline targeting the major gene families of defensins and cathelicidins to explore AMP diversity and analyze their evolution and distribution across six bat families. A combination of manual and automated procedures identified 29 AMP families across queried species, with α-, β-defensins, and cathelicidins representing around 10% of AMP diversity. Gene duplications were inferred in both α-defensins, which were absent in five species, and three β-defensin gene subfamilies, but cathelicidins did not show significant shifts in gene family size and were absent in Anoura caudifer and the pteropodids. Based on lineage-specific gains and losses, we propose diet and diet-related microbiome evolution may determine the evolution of α- and β-defensins gene families and subfamilies. These results highlight the importance of building species-specific libraries for genome annotation in non-model organisms and shed light on possible drivers responsible for the rapid evolution of AMPs. By focusing on these understudied defenses, we provide a robust framework for explaining bat responses to pathogens.
Keywords: bioinformatics pipelines; defensins; gene annotation; innate immunity; non-model organisms; transposable elements.
Copyright © 2023 Castellanos, Moreno-Santillán, Hughes, Paulat, Sipperly, Brown, Martin, Poterewicz, Lim, Russell, Moore, Johnson, Corthals, Ray and Dávalos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Delves PJ, Martin SJ, Burton DR, Roitt IM. Roitt’s essential immunology, 13th. Chichester, West Sussex: John Wiley & Sons, Ltd; (2017).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

