The Resistance and Virulence Characteristics of Salmonella Enteritidis Strain Isolated from Patients with Food Poisoning Based on the Whole-Genome Sequencing and Quantitative Proteomic Analysis
- PMID: 37823028
- PMCID: PMC10564084
- DOI: 10.2147/IDR.S411125
The Resistance and Virulence Characteristics of Salmonella Enteritidis Strain Isolated from Patients with Food Poisoning Based on the Whole-Genome Sequencing and Quantitative Proteomic Analysis
Abstract
Objective: This paper explores the drug resistance, genome and proteome expression characteristics of Salmonella from a food poisoning event.
Methods: A multidrug-resistant Salmonella Enteritidis strain, labeled as 27A, was isolated and identified from a food poisoning patient. Antimicrobial susceptibility testing determined the resistance of 27A strain to 14 antibiotics. Then, WGS analysis and comparative genomics analysis were performed on 27A, and the functional annotation of resistance genes, virulence genes were performed based on VFDB, ARDB, COG, CARD, GO, KEGG, and CAZY databases. Meanwhile, based on iTRAQ technology, quantitative proteomic analysis was conducted on 27A to analyze the functions and interactions of differentially expressed proteins related to bacterial resistance and pathogenicity.
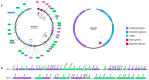
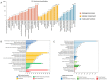
Results: Strain 27A belonged to ST11 S. Enteritidis and was resistant to levofloxacin, ciprofloxacin, ampicillin, piperacillin, and ampicillin/sulbactam. There were 33 drug resistance genes, 384 virulence genes and 2 plasmid replicon, IncFIB(S) and IncFII(S), annotated by WGS. Proteomic analysis revealed significant changes in virulence and drug proteins, which were mainly involved in bacterial pathogenicity and metabolic processes. PPI prediction showed the relationship between virulence proteins and T3SS proteins, and PagN cooperated with proteins related to T3SS to jointly mediate the invasion of 27A strain on the human body. Phylogenetic analysis indicated that S. Enteritidis has potential transmission in humans, food, and animals.
Conclusion: This study comprehensively analyzed the drug resistance and virulence phenotypes of S. Enteritidis 27A using genomic and proteomic approaches. These helps reveal the drug resistance and virulence mechanisms of S. Enteritidis, and provides important information for the source tracing and the prevention of related diseases, which lays a foundation for research on food safety, public health monitoring, and the drug resistance and pathogenicity of S. Enteritidis.
Keywords: Salmonella; WGS; evolution; quantitative proteomics; resistance; virulence.
© 2023 Xu et al.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures








References
LinkOut - more resources
Full Text Sources

