The ancestral chromatin landscape of land plants
- PMID: 37823324
- PMCID: PMC10952607
- DOI: 10.1111/nph.19311
The ancestral chromatin landscape of land plants
Abstract
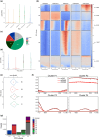
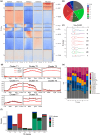
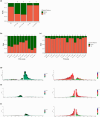
Recent studies have shown that correlations between chromatin modifications and transcription vary among eukaryotes. This is the case for marked differences between the chromatin of the moss Physcomitrium patens and the liverwort Marchantia polymorpha. Mosses and liverworts diverged from hornworts, altogether forming the lineage of bryophytes that shared a common ancestor with land plants. We aimed to describe chromatin in hornworts to establish synapomorphies across bryophytes and approach a definition of the ancestral chromatin organization of land plants. We used genomic methods to define the 3D organization of chromatin and map the chromatin landscape of the model hornwort Anthoceros agrestis. We report that nearly half of the hornwort transposons were associated with facultative heterochromatin and euchromatin and formed the center of topologically associated domains delimited by protein coding genes. Transposons were scattered across autosomes, which contrasted with the dense compartments of constitutive heterochromatin surrounding the centromeres in flowering plants. Most of the features observed in hornworts are also present in liverworts or in mosses but are distinct from flowering plants. Hence, the ancestral genome of bryophytes was likely a patchwork of units of euchromatin interspersed within facultative and constitutive heterochromatin. We propose this genome organization was ancestral to land plants.
Keywords: Anthoceros agrestis; Marchantia polymorpha; bryophytes; chromatin; epigenetic; evolution.
© 2023 The Authors. New Phytologist © 2023 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures



References
-
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nature Reviews Genetics 17: 487–500. - PubMed
-
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane‐Robinson C, Kouzarides T. 2005. Spatial distribution of di‐ and tri‐methyl lysine 36 of histone H3 at active genes. The Journal of Biological Chemistry 280: 17732–17736. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

