Direct prediction of carbapenem resistance in Pseudomonas aeruginosa by whole genome sequencing and metagenomic sequencing
- PMID: 37823665
- PMCID: PMC10662344
- DOI: 10.1128/jcm.00617-23
Direct prediction of carbapenem resistance in Pseudomonas aeruginosa by whole genome sequencing and metagenomic sequencing
Abstract
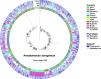
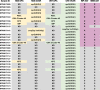
Carbapenem resistance is a major concern in the management of antibiotic-resistant Pseudomonas aeruginosa infections. The direct prediction of carbapenem-resistant phenotype from genotype in P. aeruginosa isolates and clinical samples would promote timely antibiotic therapy. The complex carbapenem resistance mechanism and the high prevalence of variant-driven carbapenem resistance in P. aeruginosa make it challenging to predict the carbapenem-resistant phenotype through the genotype. In this study, using whole genome sequencing (WGS) data of 1,622 P. aeruginosa isolates followed by machine learning, we screened 16 and 31 key gene features associated with imipenem (IPM) and meropenem (MEM) resistance in P. aeruginosa, including oprD(HIGH), and constructed the resistance prediction models. The areas under the curves of the IPM and MEM resistance prediction models were 0.906 and 0.925, respectively. For the direct prediction of carbapenem resistance in P. aeruginosa from clinical samples by the key gene features selected and prediction models constructed, 72 P. aeruginosa-positive sputum samples were collected and sequenced by metagenomic sequencing (MGS) based on next-generation sequencing (NGS) or Oxford Nanopore Technology (ONT). The prediction applicability of MGS based on NGS outperformed that of MGS based on ONT. In 72 P. aeruginosa-positive sputum samples, 65.0% (26/40) of IPM-insensitive and 63.2% (24/38) of MEM-insensitive P. aeruginosa were directly predicted by NGS-based MGS with positive predictive values of 0.897 and 0.889, respectively. By the direct detection of the key gene features associated with carbapenem resistance of P. aeruginosa, the carbapenem resistance of P. aeruginosa could be directly predicted from cultured isolates by WGS or from clinical samples by NGS-based MGS, which could assist the timely treatment and surveillance of carbapenem-resistant P. aeruginosa.
Keywords: Pseudomonas aeruginosa; carbapenem resistance prediction; machine learning; metagenomic sequencing; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Achalapong S, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Babin F-X, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Berkley JA, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Clotaire Donatien R, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, Dramowski A, Dunachie SJ, Duong Bich T, Eckmanns T, Eibach D, Emami A, Feasey N, Fisher-Pearson N, Forrest K, Garcia C, Garrett D, Gastmeier P, Giref AZ, Greer RC, Gupta V, Haller S, Haselbeck A, Hay SI, Holm M, Hopkins S, Hsia Y, Iregbu KC, Jacobs J, Jarovsky D, Javanmardi F, Jenney AWJ, Khorana M, Khusuwan S, Kissoon N, Kobeissi E, Kostyanev T, Krapp F, Krumkamp R, Kumar A, Kyu HH, Lim C, Lim K, Limmathurotsakul D, Loftus MJ, Lunn M, Ma J, Manoharan A, Marks F, May J, Mayxay M, Mturi N, Munera-Huertas T, Musicha P, Musila LA, Mussi-Pinhata MM, Naidu RN, Nakamura T, Nanavati R, Nangia S, Newton P, Ngoun C, Novotney A, Nwakanma D, Obiero CW, Ochoa TJ, Olivas-Martinez A, Olliaro P, Ooko E, Ortiz-Brizuela E, Ounchanum P, Pak GD, Paredes JL, Peleg AY, Perrone C, Phe T, Phommasone K, Plakkal N, Ponce-de-Leon A, Raad M, Ramdin T, Rattanavong S, Riddell A, Roberts T, Robotham JV, Roca A, Rosenthal VD, Rudd KE, Russell N, Sader HS, Saengchan W, Schnall J, Scott JAG, Seekaew S, Sharland M, Shivamallappa M, Sifuentes-Osornio J, Simpson AJ, Steenkeste N, Stewardson AJ, Stoeva T, Tasak N, Thaiprakong A, Thwaites G, Tigoi C, Turner C, Turner P, van Doorn HR, Velaphi S, Vongpradith A, Vongsouvath M, Vu H, Walsh T, Walson JL, Waner S, Wangrangsimakul T, Wannapinij P, Wozniak T, Young Sharma TEMW, Yu KC, Zheng P, Sartorius B, Lopez AD, Stergachis A, Moore C, Dolecek C, Naghavi M. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. doi:10.1016/S0140-6736(21)02724-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2017YFC1309700, 2017YFC1309701/MOST | National Key Research and Development Program of China (NKPs)
- shslczdzk02202/Shanghai Municipal Key Clinical Specialty
- 2017ZZ02014/Shanghai Top-Priority Clinical Key Disciplines Construction Project
- 20dz2261100/Shanghai Key Laboratory of Emergency Prevention, Diagnosis and Treatment of Respiratory Infectious Diseases
- 20dz2210500/Cultivation Projection of Shanghai Major Infectious Disease Research Base
LinkOut - more resources
Full Text Sources

