Improving radiotherapy in immunosuppressive microenvironments by targeting complement receptor C5aR1
- PMID: 37824211
- PMCID: PMC10688992
- DOI: 10.1172/JCI168277
Improving radiotherapy in immunosuppressive microenvironments by targeting complement receptor C5aR1
Abstract
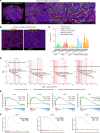
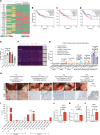
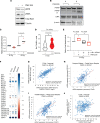
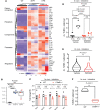
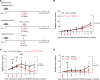
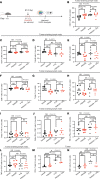
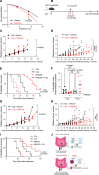
An immunosuppressive microenvironment causes poor tumor T cell infiltration and is associated with reduced patient overall survival in colorectal cancer. How to improve treatment responses in these tumors is still a challenge. Using an integrated screening approach to identify cancer-specific vulnerabilities, we identified complement receptor C5aR1 as a druggable target, which when inhibited improved radiotherapy, even in tumors displaying immunosuppressive features and poor CD8+ T cell infiltration. While C5aR1 is well-known for its role in the immune compartment, we found that C5aR1 is also robustly expressed on malignant epithelial cells, highlighting potential tumor cell-specific functions. C5aR1 targeting resulted in increased NF-κB-dependent apoptosis specifically in tumors and not normal tissues, indicating that, in malignant cells, C5aR1 primarily regulated cell fate. Collectively, these data revealed that increased complement gene expression is part of the stress response mounted by irradiated tumors and that targeting C5aR1 could improve radiotherapy, even in tumors displaying immunosuppressive features.
Keywords: Cancer; Cell Biology; Complement; Oncology; Radiation therapy.
Figures
References
-
- Llosa NJ, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.CD-14-0863. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

