Functional Pdgfra fibroblast heterogeneity in normal and fibrotic mouse lung
- PMID: 37824216
- PMCID: PMC10721331
- DOI: 10.1172/jci.insight.164380
Functional Pdgfra fibroblast heterogeneity in normal and fibrotic mouse lung
Abstract
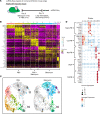
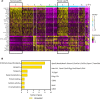
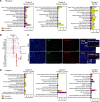
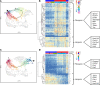
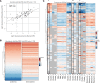
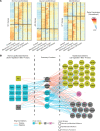
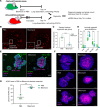
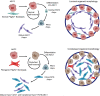
Aberrant fibroblast function plays a key role in the pathogenesis of idiopathic pulmonary fibrosis, a devastating disease of unrelenting extracellular matrix deposition in response to lung injury. Platelet-derived growth factor α-positive (Pdgfra+) lipofibroblasts (LipoFBs) are essential for lung injury response and maintenance of a functional alveolar stem cell niche. Little is known about the effects of lung injury on LipoFB function. Here, we used single-cell RNA-Seq (scRNA-Seq) technology and PdgfraGFP lineage tracing to generate a transcriptomic profile of Pdgfra+ fibroblasts in normal and injured mouse lungs 14 days after bleomycin exposure, generating 11 unique transcriptomic clusters that segregated according to treatment. While normal and injured LipoFBs shared a common gene signature, injured LipoFBs acquired fibrogenic pathway activity with an attenuation of lipogenic pathways. In a 3D organoid model, injured Pdgfra+ fibroblast-supported organoids were morphologically distinct from those cultured with normal fibroblasts, and scRNA-Seq analysis suggested distinct transcriptomic changes in alveolar epithelia supported by injured Pdgfra+ fibroblasts. In summary, while LipoFBs in injured lung have not migrated from their niche and retain their lipogenic identity, they acquire a potentially reversible fibrogenic profile, which may alter the kinetics of epithelial regeneration and potentially contribute to dysregulated repair, leading to fibrosis.
Keywords: Fibrosis; Pulmonology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

