Essential role of Mg2+ in mouse preimplantation embryo development revealed by TRPM7 chanzyme-deficient gametes
- PMID: 37824328
- PMCID: PMC10842026
- DOI: 10.1016/j.celrep.2023.113232
Essential role of Mg2+ in mouse preimplantation embryo development revealed by TRPM7 chanzyme-deficient gametes
Abstract
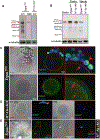
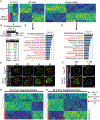
TRPM7 (transient receptor potential cation channel subfamily M member 7) is a chanzyme with channel and kinase domains essential for embryo development. Using gamete-specific Trpm7-null lines, we report that TRPM7-mediated Mg2+ influx is indispensable for reaching the blastocyst stage. TRPM7 is expressed dynamically from gametes to blastocysts; displays stage-specific localization on the plasma membrane, cytoplasm, and nucleus; and undergoes cleavage that produces C-terminal kinase fragments. TRPM7 underpins Mg2+ homeostasis, and excess Mg2+ but not Zn2+ or Ca2+ overcomes the arrest of Trpm7-null embryos; expressing Trpm7 mRNA restores development, but mutant versions fail or are partially rescued. Transcriptomic analyses of Trpm7-null embryos reveal an abundance of oxidative stress-pathway genes, confirmed by mitochondrial dysfunction, and a reduction in transcription factor networks essential for proliferation; Mg2+ supplementation corrects these defects. Hence, TRPM7 underpins Mg2+ homeostasis in preimplantation embryos, prevents oxidative stress, and promotes gene expression patterns necessary for developmental progression and cell-lineage specification.
Keywords: ART; CP: Cell biology; CP: Developmental biology; Ca(2+) oscillations; Zn(2+); eggs; fertilization; mammals; nuclear localization; oocytes; oxidative stress; sperm.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Stricker SA (1999). Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev. Biol 211, 157–176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

