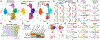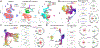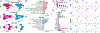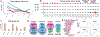Single-cell analysis of prenatal and postnatal human cortical development
- PMID: 37824647
- PMCID: PMC11005279
- DOI: 10.1126/science.adf0834
Single-cell analysis of prenatal and postnatal human cortical development
Abstract
We analyzed >700,000 single-nucleus RNA sequencing profiles from 106 donors during prenatal and postnatal developmental stages and identified lineage-specific programs that underlie the development of specific subtypes of excitatory cortical neurons, interneurons, glial cell types, and brain vasculature. By leveraging single-nucleus chromatin accessibility data, we delineated enhancer gene regulatory networks and transcription factors that control commitment of specific cortical lineages. By intersecting our results with genetic risk factors for human brain diseases, we identified the cortical cell types and lineages most vulnerable to genetic insults of different brain disorders, especially autism. We find that lineage-specific gene expression programs up-regulated in female cells are especially enriched for the genetic risk factors of autism. Our study captures the molecular progression of cortical lineages across human development.
Conflict of interest statement
Figures