Rare de novo gain-of-function missense variants in DOT1L are associated with developmental delay and congenital anomalies
- PMID: 37827158
- PMCID: PMC10645550
- DOI: 10.1016/j.ajhg.2023.09.009
Rare de novo gain-of-function missense variants in DOT1L are associated with developmental delay and congenital anomalies
Abstract
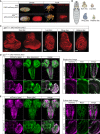
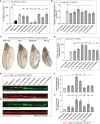
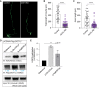
Misregulation of histone lysine methylation is associated with several human cancers and with human developmental disorders. DOT1L is an evolutionarily conserved gene encoding a lysine methyltransferase (KMT) that methylates histone 3 lysine-79 (H3K79) and was not previously associated with a Mendelian disease in OMIM. We have identified nine unrelated individuals with seven different de novo heterozygous missense variants in DOT1L through the Undiagnosed Disease Network (UDN), the SickKids Complex Care genomics project, and GeneMatcher. All probands had some degree of global developmental delay/intellectual disability, and most had one or more major congenital anomalies. To assess the pathogenicity of the DOT1L variants, functional studies were performed in Drosophila and human cells. The fruit fly DOT1L ortholog, grappa, is expressed in most cells including neurons in the central nervous system. The identified DOT1L variants behave as gain-of-function alleles in flies and lead to increased H3K79 methylation levels in flies and human cells. Our results show that human DOT1L and fly grappa are required for proper development and that de novo heterozygous variants in DOT1L are associated with a Mendelian disease.
Keywords: DOT1 Like histone lysine methyltransferase; DOT1L; Drosophila; H3K79 methylation; congenital anomalies; developmental delay; gain of function; gpp; grappa; histone lysine methyltransferase.
Copyright © 2023 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., et al. Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

