DICER ribonuclease removes harmful R-loops
- PMID: 37827159
- PMCID: PMC11034902
- DOI: 10.1016/j.molcel.2023.09.021
DICER ribonuclease removes harmful R-loops
Abstract
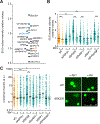
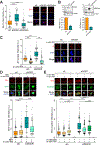
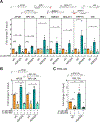
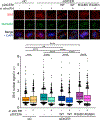
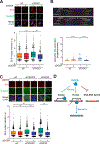
R-loops, which consist of a DNA-RNA hybrid and a displaced DNA strand, are known to threaten genome integrity. To counteract this, different mechanisms suppress R-loop accumulation by either preventing the hybridization of RNA with the DNA template (RNA biogenesis factors), unwinding the hybrid (DNA-RNA helicases), or degrading the RNA moiety of the R-loop (type H ribonucleases [RNases H]). Thus far, RNases H are the only nucleases known to cleave DNA-RNA hybrids. Now, we show that the RNase DICER also resolves R-loops. Biochemical analysis reveals that DICER acts by specifically cleaving the RNA within R-loops. Importantly, a DICER RNase mutant impaired in R-loop processing causes a strong accumulation of R-loops in cells. Our results thus not only reveal a function of DICER as an R-loop resolvase independent of DROSHA but also provide evidence for the role of multi-functional RNA processing factors in the maintenance of genome integrity in higher eukaryotes.
Keywords: DICER; DICER1 syndrome; DNA damage; R-loops; RNA nuclease; genetic instability; replication fork stalling.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Gaillard H, Garcia-Muse T, and Aguilera A. (2015). Replication stress and cancer. Nat Rev Cancer 15, 276–89. - PubMed
-
- Garcia-Muse T, and Aguilera A. (2019). R Loops: From Physiological to Pathological Roles. Cell 179, 604–618. - PubMed
-
- Huertas P, and Aguilera A. (2003). Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12, 711–21. - PubMed
-
- Li X, and Manley JL (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–78. - PubMed
-
- Aguilera A. (2005). mRNA processing and genomic instability. Nat Struct Mol Biol 12, 737–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

