Targeting PGLYRP1 promotes antitumor immunity while inhibiting autoimmune neuroinflammation
- PMID: 37828379
- PMCID: PMC10864036
- DOI: 10.1038/s41590-023-01645-4
Targeting PGLYRP1 promotes antitumor immunity while inhibiting autoimmune neuroinflammation
Abstract
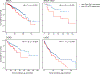
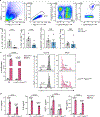
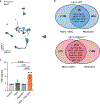
Co-inhibitory and checkpoint molecules suppress T cell function in the tumor microenvironment, thereby rendering T cells dysfunctional. Although immune checkpoint blockade is a successful treatment option for multiple human cancers, severe autoimmune-like adverse effects can limit its application. Here, we show that the gene encoding peptidoglycan recognition protein 1 (PGLYRP1) is highly coexpressed with genes encoding co-inhibitory molecules, indicating that it might be a promising target for cancer immunotherapy. Genetic deletion of Pglyrp1 in mice led to decreased tumor growth and an increased activation/effector phenotype in CD8+ T cells, suggesting an inhibitory function of PGLYRP1 in CD8+ T cells. Surprisingly, genetic deletion of Pglyrp1 protected against the development of experimental autoimmune encephalomyelitis, a model of autoimmune disease in the central nervous system. PGLYRP1-deficient myeloid cells had a defect in antigen presentation and T cell activation, indicating that PGLYRP1 might function as a proinflammatory molecule in myeloid cells during autoimmunity. These results highlight PGLYRP1 as a promising target for immunotherapy that, when targeted, elicits a potent antitumor immune response while protecting against some forms of tissue inflammation and autoimmunity.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests statement
A.C.A. is a member of the SAB for Tizona Therapeutics, Trishula Therapeutics, Compass Therapeutics, Zumutor Biologics, ImmuneOncia, and Excepgen, which have interests in cancer immunotherapy. A.C.A. is also a paid consultant for iTeos Therapeutics and Larkspur Biosciences. V.K.K. is cofounder of Celsius Therapeutics, Tizona Therapeutics, Larkspur Biosciences and Bicara Therapeutics. A.C.A.’s and V.K.K.’s interests are reviewed and managed by the Brigham and Women’s Hospital and Partners Healthcare in accordance with their conflict-of-interest policies. A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas, and was an SAB member of ThermoFisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics and Asimov until July 31, 2020. A.R. is an employee of Genentech (member of the Roche Group) since August 2020, and has equity in Roche. All other authors declare no competing interests.
Figures




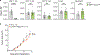





References
-
- Khan Z, Hammer C, Carroll J, Di Nucci F, Acosta SL, Maiya V, Bhangale T, Hunkapiller J, Mellman I, Albert ML, et al. (2021). Genetic variation associated with thyroid autoimmunity shapes the systemic immune response to PD-1 checkpoint blockade. Nat Commun 12, 3355. 10.1038/s41467-021-23661-4. - DOI - PMC - PubMed
Methods-only references
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

