oFVSD: a Python package of optimized forward variable selection decoder for high-dimensional neuroimaging data
- PMID: 37829329
- PMCID: PMC10566623
- DOI: 10.3389/fninf.2023.1266713
oFVSD: a Python package of optimized forward variable selection decoder for high-dimensional neuroimaging data
Abstract
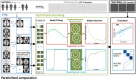
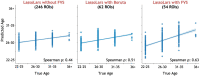
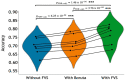
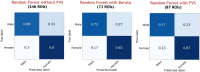
The complexity and high dimensionality of neuroimaging data pose problems for decoding information with machine learning (ML) models because the number of features is often much larger than the number of observations. Feature selection is one of the crucial steps for determining meaningful target features in decoding; however, optimizing the feature selection from such high-dimensional neuroimaging data has been challenging using conventional ML models. Here, we introduce an efficient and high-performance decoding package incorporating a forward variable selection (FVS) algorithm and hyper-parameter optimization that automatically identifies the best feature pairs for both classification and regression models, where a total of 18 ML models are implemented by default. First, the FVS algorithm evaluates the goodness-of-fit across different models using the k-fold cross-validation step that identifies the best subset of features based on a predefined criterion for each model. Next, the hyperparameters of each ML model are optimized at each forward iteration. Final outputs highlight an optimized number of selected features (brain regions of interest) for each model with its accuracy. Furthermore, the toolbox can be executed in a parallel environment for efficient computation on a typical personal computer. With the optimized forward variable selection decoder (oFVSD) pipeline, we verified the effectiveness of decoding sex classification and age range regression on 1,113 structural magnetic resonance imaging (MRI) datasets. Compared to ML models without the FVS algorithm and with the Boruta algorithm as a variable selection counterpart, we demonstrate that the oFVSD significantly outperformed across all of the ML models over the counterpart models without FVS (approximately 0.20 increase in correlation coefficient, r, with regression models and 8% increase in classification models on average) and with Boruta variable selection algorithm (approximately 0.07 improvement in regression and 4% in classification models). Furthermore, we confirmed the use of parallel computation considerably reduced the computational burden for the high-dimensional MRI data. Altogether, the oFVSD toolbox efficiently and effectively improves the performance of both classification and regression ML models, providing a use case example on MRI datasets. With its flexibility, oFVSD has the potential for many other modalities in neuroimaging. This open-source and freely available Python package makes it a valuable toolbox for research communities seeking improved decoding accuracy.
Keywords: MRI; VBM (voxel-based morphometry); forward variable selection; machine learning; neural decoding; optimized hyperparameter.
Copyright © 2023 Dang, Fermin and Machizawa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Agrawal T. (2021). “Hyperparameter optimization using Scikit-learn” in Hyperparameter optimization in machine learning (Berkeley, CA: Apress; ), 31–51.
-
- Al-Nesf M. A. Y., Abdesselem H. B., Bensmail I., Ibrahim S., Saeed W. A. H., Mohammed S. S. I., et al. (2022). Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications. Nat. Commun. 13:946. doi: 10.1038/s41467-022-28639-4, PMID: - DOI - PMC - PubMed
-
- Bergstra J., Bengio Y. (2012). Random search for hyper-parameter optimization. J. Mach. Learn. Res. 13, 281–305. doi: 10.5555/2188385.2188395 - DOI
-
- Bisong E. (2019). “More supervised machine learning techniques with Scikit-learn” in Building machine learning and deep learning models on Google cloud platform (Berkeley, United States: Apress; ), 287–308.
-
- Blanco R., Larrañaga P., Inza I., Sierra B. (2004). Gene selection for cancer classification using wrapper approaches. Int. J. Pattern Recognit. Artif. Intell. 18, 1373–1390. doi: 10.1142/S0218001404003800 - DOI
LinkOut - more resources
Full Text Sources

