Early Pharmacodynamic Changes Measured Using RNA Sequencing of Peripheral Blood from Patients in a Phase I Study with Mitazalimab, a Potent CD40 Agonistic Monoclonal Antibody
- PMID: 37830579
- PMCID: PMC10572020
- DOI: 10.3390/cells12192365
Early Pharmacodynamic Changes Measured Using RNA Sequencing of Peripheral Blood from Patients in a Phase I Study with Mitazalimab, a Potent CD40 Agonistic Monoclonal Antibody
Abstract
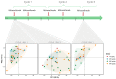
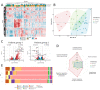
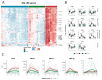
CD40-targeting therapies can enhance the dendritic cell priming of tumor-specific T cells and repolarize intratumoral macrophages to alleviate the tumoral immunosuppressive environment and remodel the extracellular matrix. Mitazalimab is a potent agonistic CD40 monoclonal IgG1 antibody currently under clinical development. This study used RNA sequencing of blood samples from a subset of patients from a Phase I trial with mitazalimab (NCT02829099) to assess peripheral pharmacodynamic activity. We found that mitazalimab induced transient peripheral transcriptomic alterations (at 600 µg/kg and 900 µg/kg dose administered intravenously), which mainly were attributed to immune activation. In particular, the transcriptomic alterations showed a reduction in effector cells (e.g., CD8+ T cells and natural killer cells) and B cells peripherally with the remaining cells (e.g., dendritic cells, monocytes, B cells, and natural killer cells) showing transcription profiles consistent with activation. Lastly, distinct patient subgroups based on the pattern of transcriptomic alterations could be identified. In summary, the data presented herein reinforce the anticipated mode of action of mitazalimab and support its ongoing clinical development.
Keywords: CD40; RNA sequencing; cancer; mitazalimab; pharmacodynamics.
Conflict of interest statement
At the time of submission, all authors are employed by, or affiliated with, Alligator Bioscience AB.
Figures

References
-
- Li F., Li C., Cai X., Xie Z., Zhou L., Cheng B., Zhong R., Xiong S., Li J., Chen Z., et al. The Association between CD8+ Tumor-Infiltrating Lymphocytes and the Clinical Outcome of Cancer Immunotherapy: A Systematic Review and Meta-Analysis. eClinicalMedicine. 2021;41:101134. doi: 10.1016/j.eclinm.2021.101134. - DOI - PMC - PubMed
-
- Liot S., Balas J., Aubert A., Prigent L., Mercier-Gouy P., Verrier B., Bertolino P., Hennino A., Valcourt U., Lambert E. Stroma Involvement in Pancreatic Ductal Adenocarcinoma: An Overview Focusing on Extracellular Matrix Proteins. Front. Immunol. 2021;12:612271. doi: 10.3389/fimmu.2021.612271. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials

