Role of Ubiquitination and Epigenetics in the Regulation of AhR Signaling in Carcinogenesis and Metastasis: "Albatross around the Neck" or "Blessing in Disguise"
- PMID: 37830596
- PMCID: PMC10571945
- DOI: 10.3390/cells12192382
Role of Ubiquitination and Epigenetics in the Regulation of AhR Signaling in Carcinogenesis and Metastasis: "Albatross around the Neck" or "Blessing in Disguise"
Abstract
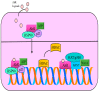
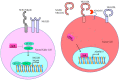
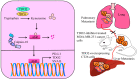
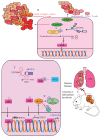
The molecular mechanisms and signal transduction cascades evoked by the activation of aryl hydrocarbon receptor (AhR) are becoming increasingly understandable. AhR is a ligand-activated transcriptional factor that integrates environmental, dietary and metabolic cues for the pleiotropic regulation of a wide variety of mechanisms. AhR mediates transcriptional programming in a ligand-specific, context-specific and cell-type-specific manner. Pioneering cutting-edge research works have provided fascinating new insights into the mechanistic role of AhR-driven downstream signaling in a wide variety of cancers. AhR ligands derived from food, environmental contaminants and intestinal microbiota strategically activated AhR signaling and regulated multiple stages of cancer. Although AhR has classically been viewed and characterized as a ligand-regulated transcriptional factor, its role as a ubiquitin ligase is fascinating. Accordingly, recent evidence has paradigmatically shifted our understanding and urged researchers to drill down deep into these novel and clinically valuable facets of AhR biology. Our rapidly increasing realization related to AhR-mediated regulation of the ubiquitination and proteasomal degradation of different proteins has started to scratch the surface of intriguing mechanisms. Furthermore, AhR and epigenome dynamics have shown previously unprecedented complexity during multiple stages of cancer progression. AhR not only transcriptionally regulated epigenetic-associated molecules, but also worked with epigenetic-modifying enzymes during cancer progression. In this review, we have summarized the findings obtained not only from cell-culture studies, but also from animal models. Different clinical trials are currently being conducted using AhR inhibitors and PD-1 inhibitors (Pembrolizumab and nivolumab), which confirm the linchpin role of AhR-related mechanistic details in cancer progression. Therefore, further studies are required to develop a better comprehension of the many-sided and "diametrically opposed" roles of AhR in the regulation of carcinogenesis and metastatic spread of cancer cells to the secondary organs.
Keywords: apoptosis; cancer; preclinical trials; signaling; xenografted mice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Rannug A., Rannug U., Rosenkranz H.S., Winqvist L., Westerholm R., Agurell E., Grafström A.K. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 1987;262:15422–15427. doi: 10.1016/S0021-9258(18)47743-5. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

