Impact of the Physical Cellular Microenvironment on the Structure and Function of a Model Hepatocyte Cell Line for Drug Toxicity Applications
- PMID: 37830622
- PMCID: PMC10572302
- DOI: 10.3390/cells12192408
Impact of the Physical Cellular Microenvironment on the Structure and Function of a Model Hepatocyte Cell Line for Drug Toxicity Applications
Abstract
It is widely recognised that cells respond to their microenvironment, which has implications for cell culture practices. Growth cues provided by 2D cell culture substrates are far removed from native 3D tissue structure in vivo. Geometry is one of many factors that differs between in vitro culture and in vivo cellular environments. Cultured cells are far removed from their native counterparts and lose some of their predictive capability and reliability. In this study, we examine the cellular processes that occur when a cell is cultured on 2D or 3D surfaces for a short period of 8 days prior to its use in functional assays, which we term: "priming". We follow the process of mechanotransduction from cytoskeletal alterations, to changes to nuclear structure, leading to alterations in gene expression, protein expression and improved functional capabilities. In this study, we utilise HepG2 cells as a hepatocyte model cell line, due to their robustness for drug toxicity screening. Here, we demonstrate enhanced functionality and improved drug toxicity profiles that better reflect the in vivo clinical response. However, findings more broadly reflect in vitro cell culture practises across many areas of cell biology, demonstrating the fundamental impact of mechanotransduction in bioengineering and cell biology.
Keywords: 3D cell culture; bioengineering; cytoskeleton; cytotoxicity; functionality; hepatocyte; mechanotransduction; microenvironment.
Conflict of interest statement
S.P. is affiliated with the company Reprocell Europe. The remaining authors declare that there are no conflict of interest to disclose.
Figures


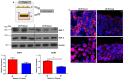



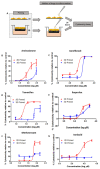

References
-
- De Volder R., Kong H. In: Biomaterials for Studies in Cellular Mechanotransduction BT—Mechanobiology of Cell-Cell and Cell-Matrix Interactions. Wagoner Johnson A., Harley B.A.C., editors. Springer; Boston, MA, USA: 2011. pp. 267–277. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

