The cystic fibrosis treatment Trikafta affects the growth, viability, and cell wall of Aspergillus fumigatus biofilms
- PMID: 37830825
- PMCID: PMC10653927
- DOI: 10.1128/mbio.01516-23
The cystic fibrosis treatment Trikafta affects the growth, viability, and cell wall of Aspergillus fumigatus biofilms
Abstract
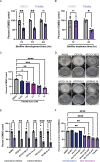
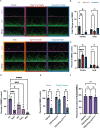
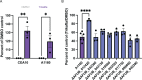
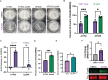
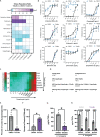
PwCF commonly test positive for pathogenic fungi, and more than 90% of the cystic fibrosis patient population is approved for the modulator treatment, Trikafta. Therefore, it is critical to understand how fungal communities, specifically A. fumigatus, respond to Trikafta exposure. Therefore, we sought to determine whether Trikafta impacted the biology of A. fumigatus biofilms. Our data demonstrate that Trikafta reduces biomass in several laboratory strains as well as clinical strains isolated from the expectorated sputum of pwCF. Furthermore, Trikafta reduces fungal viability and the capacity of biofilms to recover following treatment. Of particular importance, Trikafta affects how A. fumigatus biofilms respond to cell wall stressors, suggesting that Trikafta modulates components of the cell wall. Since the cell wall directly affects how a host immune system will respond to and effectively neutralize pathogens, our work, demonstrating that Trikafta impacts the A. fumigatus cell wall, is potentially highly relevant to fungal-induced disease pathogenesis.
Keywords: Aspergillus fumigatus; Trikafta; anti-fungal agents; cystic fibrosis; drug susceptibility.
Conflict of interest statement
The authors declare no conflict of interest
Figures

References
-
- Heijerman HGM, McKone EF, Downey DG, Van Braeckel E, Rowe SM, Tullis E, Mall MA, Welter JJ, Ramsey BW, McKee CM, Marigowda G, Moskowitz SM, Waltz D, Sosnay PR, Simard C, Ahluwalia N, Xuan F, Zhang Y, Taylor-Cousar JL, McCoy KS, VX17-445-103 Trial Group . 2019. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet 394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8 - DOI - PMC - PubMed
-
- Middleton PG, Mall MA, Dřevínek P, Lands LC, McKone EF, Polineni D, Ramsey BW, Taylor-Cousar JL, Tullis E, Vermeulen F, Marigowda G, McKee CM, Moskowitz SM, Nair N, Savage J, Simard C, Tian S, Waltz D, Xuan F, Rowe SM, Jain R, VX17-445-102 Study Group . 2019. Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single phe508del allele. N Engl J Med 381:1809–1819. doi: 10.1056/NEJMoa1908639 - DOI - PMC - PubMed
-
- Laselva O, Bartlett C, Gunawardena TNA, Ouyang H, Eckford PDW, Moraes TJ, Bear CE, Gonska T. 2021. Rescue of multiple class II CFTR mutations by elexacaftor+ tezacaftor+ivacaftor mediated in part by the dual activities of elexacaftor as both corrector and potentiator. Eur Respir J 57:2002774. doi: 10.1183/13993003.02774-2020 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

