Deciphering neuronal deficit and protein profile changes in human brain organoids from patients with creatine transporter deficiency
- PMID: 37830910
- PMCID: PMC10575631
- DOI: 10.7554/eLife.88459
Deciphering neuronal deficit and protein profile changes in human brain organoids from patients with creatine transporter deficiency
Abstract
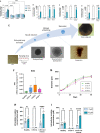
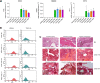
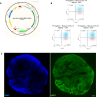
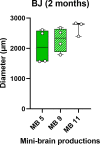
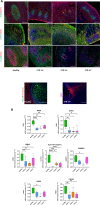
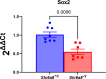
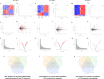
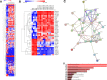
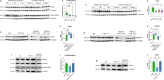
Creatine transporter deficiency (CTD) is an X-linked disease caused by mutations in the SLC6A8 gene. The impaired creatine uptake in the brain results in intellectual disability, behavioral disorders, language delay, and seizures. In this work, we generated human brain organoids from induced pluripotent stem cells of healthy subjects and CTD patients. Brain organoids from CTD donors had reduced creatine uptake compared with those from healthy donors. The expression of neural progenitor cell markers SOX2 and PAX6 was reduced in CTD-derived organoids, while GSK3β, a key regulator of neurogenesis, was up-regulated. Shotgun proteomics combined with integrative bioinformatic and statistical analysis identified changes in the abundance of proteins associated with intellectual disability, epilepsy, and autism. Re-establishment of the expression of a functional SLC6A8 in CTD-derived organoids restored creatine uptake and normalized the expression of SOX2, GSK3β, and other key proteins associated with clinical features of CTD patients. Our brain organoid model opens new avenues for further characterizing the CTD pathophysiology and supports the concept that reinstating creatine levels in patients with CTD could result in therapeutic efficacy.
Keywords: brain; cell biology; cortex; human; neurogenesis; neuroscience.
© 2023, Broca-Brisson et al.
Conflict of interest statement
LB, RH, CD, OM, RG, AM, NC, AG, BS, AA, MS, LM, FY, JA, RH, AM No competing interests declared
Figures


Update of
- doi: 10.1101/2023.06.01.543271
- doi: 10.7554/eLife.88459.1
- doi: 10.7554/eLife.88459.2
References
-
- Bruun TUJ, Sidky S, Bandeira AO, Debray F-G, Ficicioglu C, Goldstein J, Joost K, Koeberl DD, Luísa D, Nassogne M-C, O’Sullivan S, Õunap K, Schulze A, van Maldergem L, Salomons GS, Mercimek-Andrews S. Treatment outcome of creatine transporter deficiency: international retrospective cohort study. Metabolic Brain Disease. 2018;33:875–884. doi: 10.1007/s11011-018-0197-3. - DOI - PubMed
-
- Chen HR, Zhang-Brotzge X, Morozov YM, Li Y, Wang S, Zhang HH, Kuan IS, Fugate EM, Mao H, Sun YY, Rakic P, Lindquist DM, DeGrauw T, Kuan CY. Creatine transporter deficiency impairs stress adaptation and brain energetics homeostasis. JCI Insight. 2021;6:e140173. doi: 10.1172/jci.insight.140173. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

