Duplications of Human Longevity-Associated Genes Across Placental Mammals
- PMID: 37831410
- PMCID: PMC10588791
- DOI: 10.1093/gbe/evad186
Duplications of Human Longevity-Associated Genes Across Placental Mammals
Abstract
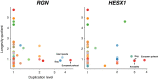
Natural selection has shaped a wide range of lifespans across mammals, with a few long-lived species showing negligible signs of ageing. Approaches used to elucidate the genetic mechanisms underlying mammalian longevity usually involve phylogenetic selection tests on candidate genes, detections of convergent amino acid changes in long-lived lineages, analyses of differential gene expression between age cohorts or species, and measurements of age-related epigenetic changes. However, the link between gene duplication and evolution of mammalian longevity has not been widely investigated. Here, we explored the association between gene duplication and mammalian lifespan by analyzing 287 human longevity-associated genes across 37 placental mammals. We estimated that the expansion rate of these genes is eight times higher than their contraction rate across these 37 species. Using phylogenetic approaches, we identified 43 genes whose duplication levels are significantly correlated with longevity quotients (False Discovery Rate (FDR) < 0.05). In particular, the strong correlation observed for four genes (CREBBP, PIK3R1, HELLS, FOXM1) appears to be driven mainly by their high duplication levels in two ageing extremists, the naked mole rat (Heterocephalus glaber) and the greater mouse-eared bat (Myotis myotis). Further sequence and expression analyses suggest that the gene PIK3R1 may have undergone a convergent duplication event, whereby the similar region of its coding sequence was independently duplicated multiple times in both of these long-lived species. Collectively, this study identified several candidate genes whose duplications may underlie the extreme longevity in mammals, and highlighted the potential role of gene duplication in the evolution of mammalian long lifespans.
Keywords: Heterocephalus glaber; Myotis myotis; gene duplication; mammalian longevity; truncated pseudogenes.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Molecular evolution of growth hormone and insulin-like growth factor 1 receptors in long-lived, small-bodied mammals.Gene. 2014 Oct 10;549(2):228-36. doi: 10.1016/j.gene.2014.07.061. Epub 2014 Jul 24. Gene. 2014. PMID: 25065922
-
Molecular adaptation of telomere associated genes in mammals.BMC Evol Biol. 2013 Nov 15;13:251. doi: 10.1186/1471-2148-13-251. BMC Evol Biol. 2013. PMID: 24237966 Free PMC article.
-
Comparative analysis of long noncoding RNAs in long-lived mammals provides insights into natural cancer-resistance.RNA Biol. 2020 Nov;17(11):1657-1665. doi: 10.1080/15476286.2020.1792116. Epub 2020 Jul 17. RNA Biol. 2020. PMID: 32635806 Free PMC article.
-
Slow aging in mammals-Lessons from African mole-rats and bats.Semin Cell Dev Biol. 2017 Oct;70:154-163. doi: 10.1016/j.semcdb.2017.07.006. Epub 2017 Jul 8. Semin Cell Dev Biol. 2017. PMID: 28698112 Review.
-
Neotenic Traits in Heterocephalus glaber and Homo sapiens.Biochemistry (Mosc). 2019 Dec;84(12):1484-1489. doi: 10.1134/S0006297919120071. Biochemistry (Mosc). 2019. PMID: 31870252 Review.
Cited by
-
Patterns of Gene Family Evolution and Selection Across Daphnia.Ecol Evol. 2025 May 24;15(5):e71453. doi: 10.1002/ece3.71453. eCollection 2025 May. Ecol Evol. 2025. PMID: 40416764 Free PMC article.
-
Different Species of Bats: Genomics, Transcriptome, and Immune Repertoire.Curr Issues Mol Biol. 2025 Apr 7;47(4):252. doi: 10.3390/cimb47040252. Curr Issues Mol Biol. 2025. PMID: 40699651 Free PMC article. Review.
-
Extensive longevity and DNA virus-driven adaptation in nearctic Myotis bats.bioRxiv [Preprint]. 2024 Nov 27:2024.10.10.617725. doi: 10.1101/2024.10.10.617725. bioRxiv. 2024. PMID: 39416019 Free PMC article. Preprint.
-
Arginines of the CGN codon family are Achilles' heels of cancer genes.Sci Rep. 2024 May 22;14(1):11715. doi: 10.1038/s41598-024-62553-7. Sci Rep. 2024. PMID: 38778164 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

