Genomic consequences associated with Agrobacterium-mediated transformation of plants
- PMID: 37831618
- PMCID: PMC10841553
- DOI: 10.1111/tpj.16496
Genomic consequences associated with Agrobacterium-mediated transformation of plants
Abstract
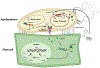
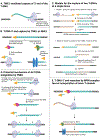
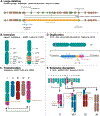
Attenuated strains of the naturally occurring plant pathogen Agrobacterium tumefaciens can transfer virtually any DNA sequence of interest to model plants and crops. This has made Agrobacterium-mediated transformation (AMT) one of the most commonly used tools in agricultural biotechnology. Understanding AMT, and its functional consequences, is of fundamental importance given that it sits at the intersection of many fundamental fields of study, including plant-microbe interactions, DNA repair/genome stability, and epigenetic regulation of gene expression. Despite extensive research and use of AMT over the last 40 years, the extent of genomic disruption associated with integrating exogenous DNA into plant genomes using this method remains underappreciated. However, new technologies like long-read sequencing make this disruption more apparent, complementing previous findings from multiple research groups that have tackled this question in the past. In this review, we cover progress on the molecular mechanisms involved in Agrobacterium-mediated DNA integration into plant genomes. We also discuss localized mutations at the site of insertion and describe the structure of these DNA insertions, which can range from single copy insertions to large concatemers, consisting of complex DNA originating from different sources. Finally, we discuss the prevalence of large-scale genomic rearrangements associated with the integration of DNA during AMT with examples. Understanding the intended and unintended effects of AMT on genome stability is critical to all plant researchers who use this methodology to generate new genetic variants.
Keywords: Agrobacterium; T-DNA concatenation; T-DNA integration; plant transformation; structural variants.
© 2023 Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
Declaration of interests
The authors declare no competing interests.
Figures

References
-
- Abdal-Aziz SA, Pliego-Alfaro F, Quesada MA and Mercado JA (2006) Evidence of frequent integration of non-T-DNA vector backbone sequences in transgenic strawberry plant. J Biosci Bioeng, 101, 508–510. - PubMed
-
- Afolabi AS, Worland B, Snape JW and Vain P. (2004) A large-scale study of rice plants transformed with different T-DNAs provides new insights into locus composition and T-DNA linkage configurations. Theor Appl Genet, 109, 815–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

