Integrin α3 promotes TH17 cell polarization and extravasation during autoimmune neuroinflammation
- PMID: 37831759
- PMCID: PMC10821720
- DOI: 10.1126/sciimmunol.adg7597
Integrin α3 promotes TH17 cell polarization and extravasation during autoimmune neuroinflammation
Abstract
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system (CNS) caused by CNS-infiltrating leukocytes, including TH17 cells that are critical mediators of disease pathogenesis. Although targeting leukocyte trafficking is effective in treating autoimmunity, there are currently no therapeutic interventions that specifically block encephalitogenic TH17 cell migration. Here, we report integrin α3 as a TH17 cell-selective determinant of pathogenicity in experimental autoimmune encephalomyelitis. CNS-infiltrating TH17 cells express high integrin α3, and its deletion in CD4+ T cells or Il17a fate-mapped cells attenuated disease severity. Mechanistically, integrin α3 enhanced the immunological synapse formation to promote the polarization and proliferation of TH17 cells. Moreover, the transmigration of TH17 cells into the CNS was dependent on integrin α3, and integrin α3 deficiency enhanced the retention of CD4+ T cells in the perivascular space of the blood-brain barrier. Integrin α3-dependent interactions continuously maintain TH17 cell identity and effector function. The requirement of integrin α3 in TH17 cell pathogenicity suggests integrin α3 as a therapeutic target for MS treatment.
Conflict of interest statement
Competing interests
The authors have no competing interests.
Figures

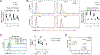
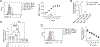





References
-
- Fallis RJ, Raine CS, McFarlin DE, Chronic relapsing experimental allergic encephalomyelitis in SJL mice following the adoptive transfer of an epitope-specific T cell line. J Neuroimmunol 22, 93–105 (1989). - PubMed
-
- Panitch HS, Hirsch RL, Haley AS, Johnson KP, Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet 1, 893–895 (1987). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

