Coinhibition of topoisomerase 1 and BRD4-mediated pause release selectively kills pancreatic cancer via readthrough transcription
- PMID: 37831776
- PMCID: PMC10575591
- DOI: 10.1126/sciadv.adg5109
Coinhibition of topoisomerase 1 and BRD4-mediated pause release selectively kills pancreatic cancer via readthrough transcription
Abstract
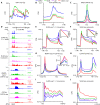
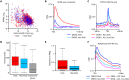
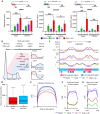
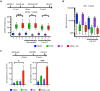
Pancreatic carcinoma lacks effective therapeutic strategies resulting in poor prognosis. Transcriptional dysregulation due to alterations in KRAS and MYC affects initiation, development, and survival of this tumor type. Using patient-derived xenografts of KRAS- and MYC-driven pancreatic carcinoma, we show that coinhibition of topoisomerase 1 (TOP1) and bromodomain-containing protein 4 (BRD4) synergistically induces tumor regression by targeting promoter pause release. Comparing the nascent transcriptome with the recruitment of elongation and termination factors, we found that coinhibition of TOP1 and BRD4 disrupts recruitment of transcription termination factors. Thus, RNA polymerases transcribe downstream of genes for hundreds of kilobases leading to readthrough transcription. This occurs during replication, perturbing replisome progression and inducing DNA damage. The synergistic effect of TOP1 + BRD4 inhibition is specific to cancer cells leaving normal cells unaffected, highlighting the tumor's vulnerability to transcriptional defects. This preclinical study provides a mechanistic understanding of the benefit of combining TOP1 and BRD4 inhibitors to treat pancreatic carcinomas addicted to oncogenic drivers of transcription and replication.
Figures


References
-
- Sur I., Taipale J., The role of enhancers in cancer. Nat. Rev. Cancer 16, 483–493 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

