A noncanonical function of SKP1 regulates the switch between autophagy and unconventional secretion
- PMID: 37831778
- PMCID: PMC10575587
- DOI: 10.1126/sciadv.adh1134
A noncanonical function of SKP1 regulates the switch between autophagy and unconventional secretion
Abstract
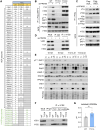
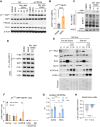
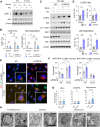
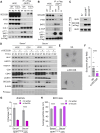
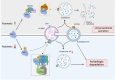
Intracellular degradation of proteins and organelles by the autophagy-lysosome system is essential for cellular quality control and energy homeostasis. Besides degradation, endolysosomal organelles can fuse with the plasma membrane and contribute to unconventional secretion. Here, we identify a function for mammalian SKP1 in endolysosomes that is independent of its established role as an essential component of the family of SCF/CRL1 ubiquitin ligases. We found that, under nutrient-poor conditions, SKP1 is phosphorylated on Thr131, allowing its interaction with V1 subunits of the vacuolar ATPase (V-ATPase). This event, in turn, promotes V-ATPase assembly to acidify late endosomes and enhance endolysosomal degradation. Under nutrient-rich conditions, SUMOylation of phosphorylated SKP1 allows its binding to and dephosphorylation by the PPM1B phosphatase. Dephosphorylated SKP1 interacts with SEC22B to promote unconventional secretion of the content of less acidified hybrid endosomal/autophagic compartments. Collectively, our study implicates SKP1 phosphorylation as a switch between autophagy and unconventional secretion in a manner dependent on cellular nutrient status.
Figures


References
-
- S. Kaushik, A. M. Cuervo, Proteostasis and aging. Nat. Med. 21, 1406–1415 (2015). - PubMed
-
- B. Levine, Autophagy and cancer. Nature 446, 745–747 (2007). - PubMed
-
- A. Fleming, M. Bourdenx, M. Fujimaki, C. Karabiyik, G. J. Krause, A. Lopez, A. Martín-Segura, C. Puri, A. Scrivo, J. Skidmore, S. M. Son, E. Stamatakou, L. Wrobel, Y. Zhu, A. M. Cuervo, D. C. Rubinsztein, The different autophagy degradation pathways and neurodegeneration. Neuron 110, 935–966 (2022). - PMC - PubMed
-
- M. D. Petroski, R. J. Deshaies, Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

