Acquired resistance to KRAS G12C small-molecule inhibitors via genetic/nongenetic mechanisms in lung cancer
- PMID: 37831779
- PMCID: PMC10575592
- DOI: 10.1126/sciadv.ade3816
Acquired resistance to KRAS G12C small-molecule inhibitors via genetic/nongenetic mechanisms in lung cancer
Abstract
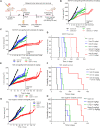
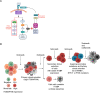
Inherent or acquired resistance to sotorasib poses a substantialt challenge for NSCLC treatment. Here, we demonstrate that acquired resistance to sotorasib in isogenic cells correlated with increased expression of integrin β4 (ITGB4), a component of the focal adhesion complex. Silencing ITGB4 in tolerant cells improved sotorasib sensitivity, while overexpressing ITGB4 enhanced tolerance to sotorasib by supporting AKT-mTOR bypass signaling. Chronic treatment with sotorasib induced WNT expression and activated the WNT/β-catenin signaling pathway. Thus, silencing both ITGB4 and β-catenin significantly improved sotorasib sensitivity in tolerant, acquired, and inherently resistant cells. In addition, the proteasome inhibitor carfilzomib (CFZ) exhibited synergism with sotorasib by down-regulating ITGB4 and β-catenin expression. Furthermore, adagrasib phenocopies the combination effect of sotorasib and CFZ by suppressing KRAS activity and inhibiting cell cycle progression in inherently resistant cells. Overall, our findings unveil previously unrecognized nongenetic mechanisms underlying resistance to sotorasib and propose a promising treatment strategy to overcome resistance.
Figures






References
-
- R. S. Herbst, D. Morgensztern, C. Boshoff, The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018). - PubMed
-
- S. Renaud, J. Seitlinger, P. E. Falcoz, M. Schaeffer, A. C. Voegeli, M. Legrain, M. Beau-Faller, G. Massard, Specific KRAS amino acid substitutions and EGFR mutations predict site-specific recurrence and metastasis following non-small-cell lung cancer surgery. Br. J. Cancer 115, 346–353 (2016). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

