Between but Not Within-Species Variation in the Distribution of Fitness Effects
- PMID: 37832225
- PMCID: PMC10630145
- DOI: 10.1093/molbev/msad228
Between but Not Within-Species Variation in the Distribution of Fitness Effects
Abstract
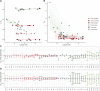
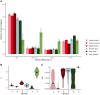
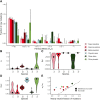
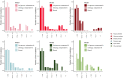
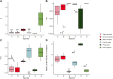
New mutations provide the raw material for evolution and adaptation. The distribution of fitness effects (DFE) describes the spectrum of effects of new mutations that can occur along a genome, and is, therefore, of vital interest in evolutionary biology. Recent work has uncovered striking similarities in the DFE between closely related species, prompting us to ask whether there is variation in the DFE among populations of the same species, or among species with different degrees of divergence, that is whether there is variation in the DFE at different levels of evolution. Using exome capture data from six tree species sampled across Europe we characterized the DFE for multiple species, and for each species, multiple populations, and investigated the factors potentially influencing the DFE, such as demography, population divergence, and genetic background. We find statistical support for the presence of variation in the DFE at the species level, even among relatively closely related species. However, we find very little difference at the population level, suggesting that differences in the DFE are primarily driven by deep features of species biology, and those evolutionarily recent events, such as demographic changes and local adaptation, have little impact.
Keywords: DFE; deleterious mutations; forest trees; population structure.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

