The NCI Imaging Data Commons as a platform for reproducible research in computational pathology
- PMID: 37832430
- PMCID: PMC10841477
- DOI: 10.1016/j.cmpb.2023.107839
The NCI Imaging Data Commons as a platform for reproducible research in computational pathology
Abstract
Background and objectives: Reproducibility is a major challenge in developing machine learning (ML)-based solutions in computational pathology (CompPath). The NCI Imaging Data Commons (IDC) provides >120 cancer image collections according to the FAIR principles and is designed to be used with cloud ML services. Here, we explore its potential to facilitate reproducibility in CompPath research.
Methods: Using the IDC, we implemented two experiments in which a representative ML-based method for classifying lung tumor tissue was trained and/or evaluated on different datasets. To assess reproducibility, the experiments were run multiple times with separate but identically configured instances of common ML services.
Results: The results of different runs of the same experiment were reproducible to a large extent. However, we observed occasional, small variations in AUC values, indicating a practical limit to reproducibility.
Conclusions: We conclude that the IDC facilitates approaching the reproducibility limit of CompPath research (i) by enabling researchers to reuse exactly the same datasets and (ii) by integrating with cloud ML services so that experiments can be run in identically configured computing environments.
Keywords: Artificial intelligence; Cloud computing; Computational pathology; FAIR; Machine learning; Reproducibility.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflicts of interest.
Figures



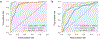
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

