Adhesion-based capture stabilizes nascent microvilli at epithelial cell junctions
- PMID: 37832537
- PMCID: PMC10615885
- DOI: 10.1016/j.devcel.2023.09.001
Adhesion-based capture stabilizes nascent microvilli at epithelial cell junctions
Abstract
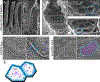
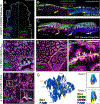
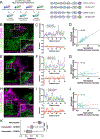
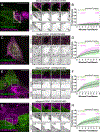
To maximize solute transport, epithelial cells build an apical "brush border," where thousands of microvilli are linked to their neighbors by protocadherin-containing intermicrovillar adhesion complexes (IMACs). Previous studies established that the IMAC is needed to build a mature brush border, but how this complex contributes to the accumulation of new microvilli during differentiation remains unclear. We found that early in differentiation, mouse, human, and porcine epithelial cells exhibit a marginal accumulation of microvilli, which span junctions and interact with protrusions on neighboring cells using IMAC protocadherins. These transjunctional IMACs are highly stable and reinforced by tension across junctions. Finally, long-term live imaging showed that the accumulation of microvilli at cell margins consistently leads to accumulation in medial regions. Thus, nascent microvilli are stabilized by a marginal capture mechanism that depends on the formation of transjunctional IMACs. These results may offer insights into how apical specializations are assembled in diverse epithelial systems.
Keywords: actin; brush border; cadherins; differentiation; intermicrovillar adhesion; transporting epithelia.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Update of
-
Adhesion-based capture stabilizes nascent microvilli at epithelial cell junctions.bioRxiv [Preprint]. 2023 Mar 9:2023.03.08.531705. doi: 10.1101/2023.03.08.531705. bioRxiv. 2023. Update in: Dev Cell. 2023 Oct 23;58(20):2048-2062.e7. doi: 10.1016/j.devcel.2023.09.001. PMID: 36945471 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

