Effect of seven anti-tuberculosis treatment regimens on sputum microbiome: a retrospective analysis of the HIGHRIF study 2 and PanACEA MAMS-TB clinical trials
- PMID: 37832571
- PMCID: PMC7617392
- DOI: 10.1016/S2666-5247(23)00191-X
Effect of seven anti-tuberculosis treatment regimens on sputum microbiome: a retrospective analysis of the HIGHRIF study 2 and PanACEA MAMS-TB clinical trials
Abstract
Background: Respiratory tract microbiota has been described as the gatekeeper for respiratory health. We aimed to assess the impact of standard-of-care and experimental anti-tuberculosis treatment regimens on the respiratory microbiome and implications for treatment outcomes.
Methods: In this retrospective study, we analysed the sputum microbiome of participants with tuberculosis treated with six experimental regimens versus standard-of-care who were part of the HIGHRIF study 2 (NCT00760149) and PanACEA MAMS-TB (NCT01785186) clinical trials across a 3-month treatment follow-up period. Samples were from participants in Mbeya, Kilimanjaro, Bagamoyo, and Dar es Salaam, Tanzania. Experimental regimens were composed of different combinations of rifampicin (R), isoniazid (H), pyrazinamide (Z), ethambutol (E), moxifloxacin (M), and a new drug, SQ109 (Q). Reverse transcription was used to create complementary DNA for each participant's total sputum RNA and the V3-V4 region of the 16S rRNA gene was sequenced using the Illumina metagenomic technique. Qiime was used to analyse the amplicon sequence variants and estimate alpha diversity. Descriptive statistics were applied to assess differences in alpha diversity pre-treatment and post-treatment initiation and the effect of each treatment regimen.
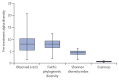
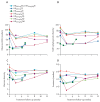
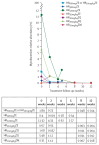
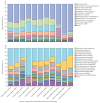
Findings: Sequence data were obtained from 397 pre-treatment and post-treatment samples taken between Sept 26, 2008, and June 30, 2015, across seven treatment regimens. Pre-treatment microbiome (206 genera) was dominated by Firmicutes (2860 [44%] of 6500 amplicon sequence variants [ASVs]) at the phylum level and Streptococcus (2340 [36%] ASVs) at the genus level. Two regimens had a significant depressing effect on the microbiome after 2 weeks of treatment, HR20mg/kgZM (Shannon diversity index p=0·0041) and HR35mg/kgZE (p=0·027). Gram-negative bacteria were the most sensitive to bactericidal activity of treatment with the highest number of species suppressed being under the moxifloxacin regimen. By week 12 after treatment initiation, microbiomes had recovered to pre-treatment level except for the HR35mg/kgZE regimen and for genus Mycobacterium, which did not show recovery across all regimens. Tuberculosis culture conversion to negative by week 8 of treatment was associated with clearance of genus Neisseria, with a 98% reduction of the pre-treatment level.
Interpretation: HR20mg/kgZM was effective against tuberculosis without limiting microbiome recovery, which implies a shorter efficacious anti-tuberculosis regimen with improved treatment outcomes might be achieved without harming the commensal microbiota.
Funding: European and Developing Countries Clinical Trials Partnership and German Ministry of Education and Research.
Copyright © 2023 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests WS and SHG provide pro bono advice for a company that is developing tuberculosis molecular bacterial load assay for clinical use. All other authors declare no competing interests.
Figures
References
-
- Uplekar M, Weil D, Lonnroth K, et al. WHO’s new end TB strategy. Lancet. 2015;385:1799–801. - PubMed

