The coenzyme A precursor pantethine enhances antitumor immunity in sarcoma
- PMID: 37833072
- PMCID: PMC10583838
- DOI: 10.26508/lsa.202302200
The coenzyme A precursor pantethine enhances antitumor immunity in sarcoma
Erratum in
-
Correction: The coenzyme A precursor pantethine enhances antitumor immunity in sarcoma.Life Sci Alliance. 2023 Nov 29;7(2):e202302479. doi: 10.26508/lsa.202302479. Print 2024 Feb. Life Sci Alliance. 2023. PMID: 38030222 Free PMC article.
Abstract
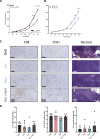
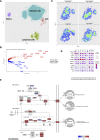
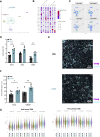
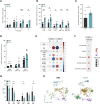
The tumor microenvironment is a dynamic network of stromal, cancer, and immune cells that interact and compete for resources. We have previously identified the Vanin1 pathway as a tumor suppressor of sarcoma development via vitamin B5 and coenzyme A regeneration. Using an aggressive sarcoma cell line that lacks Vnn1 expression, we showed that the administration of pantethine, a vitamin B5 precursor, attenuates tumor growth in immunocompetent but not nude mice. Pantethine boosts antitumor immunity, including the polarization of myeloid and dendritic cells towards enhanced IFNγ-driven antigen presentation pathways and improved the development of hypermetabolic effector CD8+ T cells endowed with potential antitumor activity. At later stages of treatment, the effect of pantethine was limited by the development of immune cell exhaustion. Nevertheless, its activity was comparable with that of anti-PD1 treatment in sensitive tumors. In humans, VNN1 expression correlates with improved survival and immune cell infiltration in soft-tissue sarcomas, but not in osteosarcomas. Pantethine could be a potential therapeutic immunoadjuvant for the development of antitumor immunity.
© 2023 Miallot et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures













References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
