Brain proteomic atlas of alcohol use disorder in adult males
- PMID: 37833300
- PMCID: PMC10575941
- DOI: 10.1038/s41398-023-02605-0
Brain proteomic atlas of alcohol use disorder in adult males
Abstract
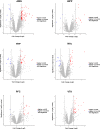
Alcohol use disorder (AUD) affects transcriptomic, epigenetic and proteomic expression in several organs, including the brain. There has not been a comprehensive analysis of altered protein abundance focusing on the multiple brain regions that undergo neuroadaptations occurring in AUD. We performed a quantitative proteomic analysis using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of human postmortem tissue from brain regions that play key roles in the development and maintenance of AUD, the amygdala (AMG), hippocampus (HIPP), hypothalamus (HYP), nucleus accumbens (NAc), prefrontal cortex (PFC) and ventral tegmental area (VTA). Brain tissues were from adult males with AUD (n = 11) and matched controls (n = 16). Across the two groups, there were >6000 proteins quantified with differential protein abundance in AUD compared to controls in each of the six brain regions. The region with the greatest number of differentially expressed proteins was the AMG, followed by the HYP. Pathways associated with differentially expressed proteins between groups (fold change > 1.5 and LIMMA p < 0.01) were analyzed by Ingenuity Pathway Analysis (IPA). In the AMG, adrenergic, opioid, oxytocin, GABA receptor and cytokine pathways were among the most enriched. In the HYP, dopaminergic signaling pathways were the most enriched. Proteins with differential abundance in AUD highlight potential therapeutic targets such as oxytocin, CSNK1D (PF-670462), GABAB receptor and opioid receptors and may lead to the identification of other potential targets. These results improve our understanding of the molecular alterations of AUD across brain regions that are associated with the development and maintenance of AUD. Proteomic data from this study is publicly available at www.lmdomics.org/AUDBrainProteomeAtlas/ .
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

