Interaction of Mesonivirus and Negevirus with arboviruses and the RNAi response in Culex tarsalis-derived cells
- PMID: 37833743
- PMCID: PMC10576325
- DOI: 10.1186/s13071-023-05985-w
Interaction of Mesonivirus and Negevirus with arboviruses and the RNAi response in Culex tarsalis-derived cells
Abstract
Background: Mosquito-specific viruses (MSVs) comprise a variety of different virus families, some of which are known to interfere with infections of medically important arboviruses. Viruses belonging to the family Mesoniviridae or taxon Negevirus harbor several insect-specific viruses, including MSVs, which are known for their wide geographical distribution and extensive host ranges. Although these viruses are regularly identified in mosquitoes all over the world, their presence in mosquitoes in Germany had not yet been reported.
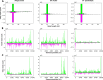
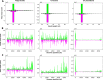
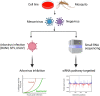
Methods: A mix of three MSVs (Yichang virus [Mesoniviridae] and two negeviruses [Daeseongdong virus and Dezidougou virus]) in a sample that contained a pool of Coquillettidia richiardii mosquitoes collected in Germany was used to investigate the interaction of these viruses with different arboviruses in Culex-derived cells. In addition, small RNA sequencing and analysis of different mosquito-derived cells infected with this MSV mix were performed.
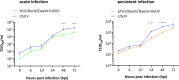
Results: A strain of Yichang virus (Mesoniviridae) and two negeviruses (Daeseongdong virus and Dezidougou virus) were identified in the Cq. richiardii mosquitoes sampled in Germany, expanding current knowledge of their circulation in central Europe. Infection of mosquito-derived cells with these three viruses revealed that they are targeted by the small interfering RNA (siRNA) pathway. In Culex-derived cells, co-infection by these three viruses had varying effects on the representative arboviruses from different virus families (Togaviridae: Semliki forest virus [SFV]; Bunyavirales: Bunyamwera orthobunyavirus [BUNV]; or Flaviviridae: Usutu virus [USUV]). Specifically, persistent MSV co-infection inhibited BUNV infection, as well as USUV infection (but the latter only at specific time points). However, the impact on SFV infection was only noticeable at low multiplicity of infection (MOI 0.1) and at specific time points in combination with the infection status.
Conclusions: Taken together, these results are important findings that will lead to a better understanding of the complex interactions of MSVs, mosquitoes and arboviruses.
Keywords: Aedes; Arbovirus; Culex; Interference; Mesonivirus; Mosquito-specific viruses; Negevirus; RNAi.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Fredericks AC, Russell TA, Wallace LE, Davidson AD, Fernandez-Sesma A, Maringer K. Aedes aegypti (Aag2)-derived clonal mosquito cell lines reveal the effects of pre-existing persistent infection with the insect-specific bunyavirus Phasi Charoen-like virus on arbovirus replication. PLoS Negl Trop Dis. 2019;13:e0007346. doi: 10.1371/journal.pntd.0007346. - DOI - PMC - PubMed
-
- Olmo RP, Todjro YMH, Aguiar ERGR, de Almeida JPP, Armache JN, de Faria IJS, et al. Insect-specific viruses regulate vector competence in Aedes aegypti mosquitoes via expression of histone H4. bioRxiv. 2021. 10.1101/2021.06.05.447047.
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

