Unusual Presentation of SET::NUP214-Associated Concomitant Hematological Neoplasm in a Child-Diagnostic and Treatment Struggle
- PMID: 37833906
- PMCID: PMC10572181
- DOI: 10.3390/ijms241914451
Unusual Presentation of SET::NUP214-Associated Concomitant Hematological Neoplasm in a Child-Diagnostic and Treatment Struggle
Abstract
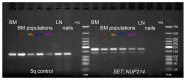
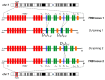
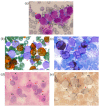
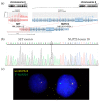
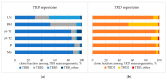
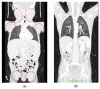
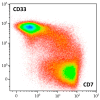
Simultaneous multilineage hematologic malignancies are uncommon and associated with poorer prognosis than single-lineage leukemia or lymphoma. Here, we describe a concomitant malignant neoplasm in a 4-year-old boy. The child presented with massive lymphoproliferative syndrome, nasal breathing difficulties, and snoring. Morphological, immunocytochemical, and flow cytometry diagnostics showed coexistence of acute myeloid leukemia (AML) and peripheral T-cell lymphoma (PTCL). Molecular examination revealed a rare t(9;9)(q34;q34)/SET::NUP214 translocation as well as common TCR clonal rearrangements in both the bone marrow and lymph nodes. The disease showed primary refractoriness to both lymphoid and myeloid high-dose chemotherapy as well as combined targeted therapy (trametinib + ruxolitinib). Hence, HSCT was performed, and the patient has since been in complete remission for over a year. This observation highlights the importance of molecular techniques for determining the united nature of complex SET::NUP214-positive malignant neoplasms arising from precursor cells with high lineage plasticity.
Keywords: HSCT; NUP214; fusion genes; pediatric leukemia and lymphoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Noone A.M., Howlader N., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. SEER Cancer Statistics Review, 1975–2015. National Cancer Institute; Bethesda, MD, USA: 2018. [(accessed on 10 January 2023)]. Available online: https://seer.cancer.gov/csr/1975_2015/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

