Foci-Xpress: Automated and Fast Nuclear Foci Counting Tool
- PMID: 37833912
- PMCID: PMC10572366
- DOI: 10.3390/ijms241914465
Foci-Xpress: Automated and Fast Nuclear Foci Counting Tool
Abstract
In the nucleus, distinct, discrete spots or regions called "foci" have been identified, each harboring a specific molecular function. Accurate and efficient quantification of these foci is essential for understanding cellular dynamics and signaling pathways. In this study, we present an innovative automated image analysis method designed to precisely quantify subcellular foci within the cell nucleus. Manual foci counting methods can be tedious and time-consuming. To address these challenges, we developed an open-source software that automatically counts the number of foci from the indicated image files. We compared the foci counting efficiency, velocity, accuracy, and convenience of Foci-Xpress with those of other conventional methods in foci-induced models. We can adjust the brightness of foci to establish a threshold. The Foci-Xpress method was significantly faster than other conventional methods. Its accuracy was similar to that of conventional methods. The most significant strength of Foci-Xpress is automation, which eliminates the need for analyzing equipment while counting. This enhanced throughput facilitates comprehensive statistical analyses and supports robust conclusions from experiments. Furthermore, automation completely rules out biases caused by researchers, such as manual errors or daily variations. Thus, Foci-Xpress is a convincing, convenient, and easily accessible focus-counting tool for cell biologists.
Keywords: DNA damage; automated screening method; foci quantitative analysis; image processing; nuclear foci.
Conflict of interest statement
We wish to confirm that the manuscript being submitted for publication has not been previously published in any scientific journal, nor is it currently under review by any other journal. Furthermore, we would like to state that none of the authors have submitted or plan to submit any similar papers, except for abstracts or preliminary communications. All authors have thoroughly reviewed and agreed upon the content of the manuscript and have provided their consent to be listed as authors. We can also confirm that no conflict of interest, including financial interests, exist among the authors with regards to the submitted manuscript.
Figures


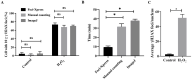

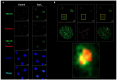
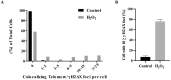
References
-
- Kruhlak M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Muller W.G., McNally J.G., Bazett-Jones D.P., Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. - DOI - PMC - PubMed
-
- Abd-Elsayed A.A., Sessler D.I., Mendoza-Cuartas M., Dalton J.E., Said T., Meinert J., Upton G., Franklin C., Kurz A. A randomized controlled study to assess patients’ understanding of and consenting for clinical trials using two different consent form presentations. Minerva Anestesiol. 2012;78:564–573. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

