Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients
- PMID: 37834231
- PMCID: PMC10573268
- DOI: 10.3390/ijms241914783
Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients
Abstract
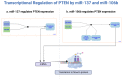
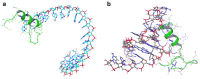
The challenge of rapidly diagnosing myocardial ischemia in unstable angina (UA) patients presenting to the Emergency Department (ED) is due to a lack of sensitive blood biomarkers. This has prompted an investigation into microRNAs (miRNAs) related to cardiac-derived Nourin for potential diagnostic application. The Nourin protein is rapidly expressed in patients with acute coronary syndrome (ACS) (UA and acute myocardial infarction (AMI)). MicroRNAs regulate gene expression through mRNA binding and, thus, may represent potential biomarkers. We initially identified miR-137 and miR-106b and conducted a clinical validation, which demonstrated that they were highly upregulated in ACS patients, but not in healthy subjects and non-ACS controls. Using integrated comprehensive bioinformatics analysis, the present study confirms that the Nourin protein targets miR-137 and miR-106b, which are linked to myocardial ischemia and inflammation associated with ACS. Molecular docking demonstrated robust interactions between the Nourin protein and miR137/hsa-miR-106b, involving hydrogen bonds and hydrophobic interactions, with -10 kcal/mol binding energy. I-TASSER generated Nourin analogs, with the top 10 chosen for structural insights. Antigenic regions and MHCII epitopes within the Nourin SPGADGNGGEAMPGG sequence showed strong binding to HLA-DR/DQ alleles. The Cytoscape network revealed interactions of -miR137/hsa-miR--106b and Phosphatase and tensin homolog (PTEN) in myocardial ischemia. RNA Composer predicted the secondary structure of miR-106b. Schrödinger software identified key Nourin-RNA interactions critical for complex stability. The study identifies miR-137 and miR-106b as potential ACS diagnostic and therapeutic targets. This research underscores the potential of miRNAs targeting Nourin for precision ACS intervention. The analysis leverages RNA Composer, Schrödinger, and I-TASSER tools to explore interactions and structural insights. Robust Nourin-miRNA interactions are established, bolstering the case for miRNA-based interventions in ischemic injury. In conclusion, the study contributes to UA and AMI diagnosis strategies through bioinformatics-guided exploration of Nourin-targeting miRNAs. Supported by comprehensive molecular analysis, the hypoxia-induced miR-137 for cell apoptosis (a marker of cell damage) and the inflammation-induced miR-106b (a marker of inflammation) confirmed their potential clinical use as diagnostic biomarkers. This research reinforces the growing role of miR-137/hsa-miR-106b in the early diagnosis of myocardial ischemia in unstable angina patients.
Keywords: Nourin-dependent miR-137 and miR-106b; integrated bioinformatics analysis; molecular dynamics; myocardial ischemia; unstable angina.
Conflict of interest statement
S.A. Elgebaly is the founder of Nour Heart, Inc. This research work is independent and objective. The other authors have no conflicts of interest to disclose.
Figures





References
-
- Roth G.A., Mensah G.A., Johnson C.O., Addolorato G., Ammirati E., Baddour L.M., Barengo N.C., Beaton A.Z., Benjamin E.J., Benziger C.P., et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020;76:2982–3021. doi: 10.1016/j.jacc.2020.11.010. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

