Modeling the Homologous Recombination Process: Methods, Successes and Challenges
- PMID: 37834348
- PMCID: PMC10573387
- DOI: 10.3390/ijms241914896
Modeling the Homologous Recombination Process: Methods, Successes and Challenges
Abstract
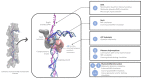
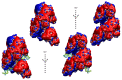
Homologous recombination (HR) is a fundamental process common to all species. HR aims to faithfully repair DNA double strand breaks. HR involves the formation of nucleoprotein filaments on DNA single strands (ssDNA) resected from the break. The nucleoprotein filaments search for homologous regions in the genome and promote strand exchange with the ssDNA homologous region in an unbroken copy of the genome. HR has been the object of intensive studies for decades. Because multi-scale dynamics is a fundamental aspect of this process, studying HR is highly challenging, both experimentally and using computational approaches. Nevertheless, knowledge has built up over the years and has recently progressed at an accelerated pace, borne by increasingly focused investigations using new techniques such as single molecule approaches. Linking this knowledge to the atomic structure of the nucleoprotein filament systems and the succession of unstable, transient intermediate steps that takes place during the HR process remains a challenge; modeling retains a very strong role in bridging the gap between structures that are stable enough to be observed and in exploring transition paths between these structures. However, working on ever-changing long filament systems submitted to kinetic processes is full of pitfalls. This review presents the modeling tools that are used in such studies, their possibilities and limitations, and reviews the advances in the knowledge of the HR process that have been obtained through modeling. Notably, we will emphasize how cooperative behavior in the HR nucleoprotein filament enables modeling to produce reliable information.
Keywords: DNA stretching; RecA; homologous recombination; integrative modeling; molecular dynamics simulations; molecular modeling; multi-scale dynamics; protein filament; protein-DNA interaction.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Figures



References
-
- Bianco P.R., Tracy R.B., Kowalczykowski S.C. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

