Phenotypic and Genotypic Characteristics of Non-Hemolytic L. monocytogenes Isolated from Food and Processing Environments
- PMID: 37835283
- PMCID: PMC10572806
- DOI: 10.3390/foods12193630
Phenotypic and Genotypic Characteristics of Non-Hemolytic L. monocytogenes Isolated from Food and Processing Environments
Abstract
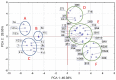
Increasingly, Listeria monocytogenes (LM) with atypical phenotypic and genotypic characteristics are being isolated from food, causing problems with their classification and testing. From 2495 soil, food, and swab samples from the food industry, 262 LM isolates were found. A total of 30 isolates were isolated, mainly from soil and plant food, and were classified as atypical LM (aLM) because they lacked the ability to move (30/11.4%) and perform hemolysis (25/9.5%). The isolation environment affected aLM incidence, cell size, sugar fermentation capacity, antibiotic sensitivity, and the number of virulence genes. Therefore, despite several characteristics differentiating all aLMs/non-hemolytic isolates from reference LMs, the remaining phenotypic characteristics were specific to each aLM isolate (like a fingerprint). The aLM/non-hemolytic isolates, particularly those from the soil and meat industries, showed more variability in their sugar fermentation capacity and were less sensitive to antibiotics than LMs. As many as 11 (36.7%) aLM isolates had resistance to four different antibiotics or simultaneously to two antibiotics. The aLM isolates possessed 3-7 of the 12 virulence genes: prfA and hly in all aLMs, while iap was not present. Only five (16.7%) isolates were classified into serogroups 1/2c-3c or 4a-4c. The aLM/non-hemolytic isolates differed by many traits from L. immobilis and atypical L. innocua. The reference method of reviving and isolating LM required optimization of aLM. Statistical analyses of clustering, correlation, and PCA showed similarities and differences between LM and aLM/non-hemolytic isolates due to individual phenotypic traits and genes. Correlations were found between biochemical traits, antibiotic resistance, and virulence genes. The increase in the incidence of atypical non-hemolytic LM may pose a risk to humans, as they may not be detected by ISO methods and have greater antibiotic resistance than LM. aLM from LM can be distinguished based on lack of hemolysis, motility, growth at 4 °C, ability to ferment D-arabitol, and lack of six specific genes.
Keywords: antibiotic resistance; atypical Listeria monocytogenes; phenotyping; serotyping; sugar fermentation; virulence genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- CDC Listeria Outbreaks, Centers for Disease Control and Prevention. [(accessed on 15 February 2023)];2023 Available online: https://www.cdc.gov/listeria/outbreaks/index.html.
-
- Carlin C.R., Liao J., Weller D., Guo X., Orsi R., Wiedmann M. Listeria cossartiae sp. nov., Listeria immobilis sp. nov., Listeria portnoyi sp. nov. and Listeria rustica sp. nov., isolated from agricultural water and natural environments. Int. J. Syst. Evol. Microbiol. 2021;71:004795. doi: 10.1099/ijsem.0.004795. - DOI - PMC - PubMed
-
- Den Bakker H.C., Warchocki S., Wright E.M., Allred A.F., Ahlstrom C., Manuel C.S., Stasiewicz M.J., Burrell A., Roof S., Strawn L.K., et al. Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov and Listeria grandensis sp. nov., from agricultural and natural environments. Int. J. Syst. Evol. Microbiol. 2014;64:1882–1889. doi: 10.1099/ijs.0.052720-0. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

