High-Dimensional Mass Cytometry Analysis of Embryonic Antigens and Their Signaling Pathways in Myeloid Cells from Bone Marrow Aspirates in AML Patients at Diagnosis
- PMID: 37835401
- PMCID: PMC10571794
- DOI: 10.3390/cancers15194707
High-Dimensional Mass Cytometry Analysis of Embryonic Antigens and Their Signaling Pathways in Myeloid Cells from Bone Marrow Aspirates in AML Patients at Diagnosis
Abstract
Background: Embryonic antigens (EA) regulate pluripotency, self-renewal, and differentiation in embryonic stem (ES) cells during their development. In adult somatic cells, EA expression is normally inhibited; however, EAs can be re-expressed by cancer cells and are involved in the deregulation of different signaling pathways (SPs). In the context of AML, data concerning the expression of EAs are scarce and contradictory.
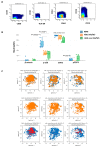
Methods: We used mass cytometry to explore the expression of EAs and three SPs in myeloid cells from AML patients and normal bone marrow (NBM). Imaging flow cytometry was used for morphological assessment of cells in association with their OCT3/4 expression status (positive vs. negative).
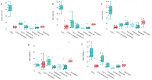
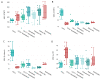
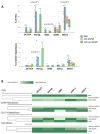
Results: An overall reduction in or absence of EA expression was observed in immature myeloid cells from AML patients compared to their normal counterparts. Stage-specific embryonic antigen-3 (SSEA-3) was consistently expressed at low levels in immature myeloid cells, whereas SSEA-1 was overexpressed in hematopoietic stem cells (HSCs) and myeloblasts from AML with monocytic differentiation (AML M4/M5). Therefore, these markers are valuable for distinguishing between normal and abnormal myeloid cells. These preliminary results show that the exploration of myeloid cell intracellular SPs in the setting of AML is very informative. Deregulation of three important leukemogenic SPs was also observed in myeloid cells from AML.
Conclusions: Exploring EAs and SPs in myeloid cells from AML patients by mass cytometry may help identify characteristic phenotypes and facilitate AML follow-up.
Keywords: Nanog; OCT3/4; SOX2; SSEA1; SSEA3; STAT3/p-STAT3; acute myeloid leukemia; mass cytometry; p-p38; β-catenin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Immunoreactivity of MIC2 (CD99) and terminal deoxynucleotidyl transferase in bone marrow clot and core specimens of acute myeloid leukemias and myelodysplastic syndromes.Arch Pathol Lab Med. 2006 Feb;130(2):153-7. doi: 10.5858/2006-130-153-IOMCAT. Arch Pathol Lab Med. 2006. PMID: 16454553
-
The value of CD64 expression in distinguishing acute myeloid leukemia with monocytic differentiation from other subtypes of acute myeloid leukemia: a flow cytometric analysis of 64 cases.Arch Pathol Lab Med. 2007 May;131(5):748-54. doi: 10.5858/2007-131-748-TVOCEI. Arch Pathol Lab Med. 2007. PMID: 17488160
-
Expression of embryonic stem cell markers in acute myeloid leukemia.Tumour Biol. 2017 Jul;39(7):1010428317716629. doi: 10.1177/1010428317716629. Tumour Biol. 2017. PMID: 28718379
-
Acute myeloid leukemias expressing lymphoid-associated antigens: diagnostic incidence and prognostic significance.Leukemia. 1993 Apr;7(4):489-98. Leukemia. 1993. PMID: 7681917 Review.
-
[Biological properties and sensitivity to induction therapy of differentiated cells expressing atypical immunophenotype in acute leukemia of children].Folia Med Cracov. 2001;42(3):5-80. Folia Med Cracov. 2001. PMID: 12353422 Review. Polish.
References
-
- Kashyap V., Rezende N.C., Scotland K.B., Shaffer S.M., Persson J.L., Gudas L.J., Mongan N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. - DOI - PMC - PubMed
-
- Qinjun Z., Hongying R., Sizhou F., Ying C., Yi H., Donglin Y., Fengxia M., Jianping L., Shihong L., Fang C., et al. Aberrant expression and significance of OCT-4A transcription factor in leukemia cells. Blood Cells Mol. Dis. 2015;54:90–96. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

