A Network of 17 Microtubule-Related Genes Highlights Functional Deregulations in Breast Cancer
- PMID: 37835564
- PMCID: PMC10571893
- DOI: 10.3390/cancers15194870
A Network of 17 Microtubule-Related Genes Highlights Functional Deregulations in Breast Cancer
Abstract
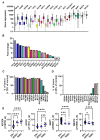
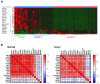
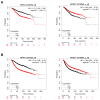
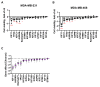
A wide panel of microtubule-associated proteins and kinases is involved in coordinated regulation of the microtubule cytoskeleton and may thus represent valuable molecular markers contributing to major cellular pathways deregulated in cancer. We previously identified a panel of 17 microtubule-related (MT-Rel) genes that are differentially expressed in breast tumors showing resistance to taxane-based chemotherapy. In the present study, we evaluated the expression, prognostic value and functional impact of these genes in breast cancer. We show that 14 MT-Rel genes (KIF4A, ASPM, KIF20A, KIF14, TPX2, KIF18B, KIFC1, AURKB, KIF2C, GTSE1, KIF15, KIF11, RACGAP1, STMN1) are up-regulated in breast tumors compared with adjacent normal tissue. Six of them (KIF4A, ASPM, KIF20A, KIF14, TPX2, KIF18B) are overexpressed by more than 10-fold in tumor samples and four of them (KIF11, AURKB, TPX2 and KIFC1) are essential for cell survival. Overexpression of all 14 genes, and underexpression of 3 other MT-Rel genes (MAST4, MAPT and MTUS1) are associated with poor breast cancer patient survival. A Systems Biology approach highlighted three major functional networks connecting the 17 MT-Rel genes and their partners, which are centered on spindle assembly, chromosome segregation and cytokinesis. Our studies identified mitotic Aurora kinases and their substrates as major targets for therapeutic approaches against breast cancer.
Keywords: Aurora kinases; Systems Biology; biomarker; kinesins; mitotic defects; prognostic value; therapeutic targets.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

