Ex Vivo Colonic Fermentation of NUTRIOSE® Exerts Immuno-Modulatory Properties and Strong Anti-Inflammatory Effects
- PMID: 37836513
- PMCID: PMC10574048
- DOI: 10.3390/nu15194229
Ex Vivo Colonic Fermentation of NUTRIOSE® Exerts Immuno-Modulatory Properties and Strong Anti-Inflammatory Effects
Abstract
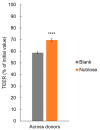
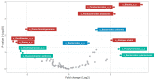
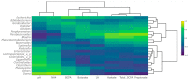
NUTRIOSE® (Roquette, Lestrem, France) is a resistant dextrin with well-established prebiotic effects. This study evaluated the indirect effects of pre-digested NUTRIOSE® on host immune response and gut barrier integrity. Fecal samples from eight healthy donors were inoculated in a Colon-on-a-plate® system (ProDigest, Ghent, Belgium) with or without NUTRIOSE® supplementation. Following 48 h fermentation, colonic suspensions were tested in a Caco-2/THP1-Blue™ co-culture system to determine their effects on gut barrier activity (transepithelial electrical resistance) and immune response following lipopolysaccharide stimulation. Additionally, changes in short-chain fatty acid levels (SCFA) and microbial community composition following a 48 h fermentation in the Colon-on-a-plate® system were measured. Across all donors, immune-mediated intestinal barrier damage was significantly reduced with NUTRIOSE®-supplemented colonic suspensions versus blank. Additionally, IL-6 and IL-10 levels were significantly increased, and the level of the neutrophil chemoattractant IL-8 was significantly decreased with NUTRIOSE®-supplemented colonic suspensions versus blank in the co-culture models following lipopolysaccharide stimulation. These beneficial effects of NUTRIOSE® supplementation were likely due to increased acetate and propionate levels and the enrichment of SCFA-producing bacteria. NUTRIOSE® was well fermented by the colonic bacteria of all eight donors and had protective effects on inflammation-induced disruption of the intestinal epithelial barrier and strong anti-inflammatory effects.
Keywords: Colon-on-a-plate; anti-inflammatory; immunomodulation; prebiotic; resistant dextrin; short-chain fatty acid.
Conflict of interest statement
Authors are employees of Roquette (C.P. and C.T.) or ProDigest (L.V., J.G., and M.M.) as indicated by our affiliations. This work and the article processing charge were funded by Roquette. ProDigest is a company based in Ghent, Belgium which specializes in offering pre-clinical services to food and functional food companies. The funders were involved in the design of the study, in the writing of the manuscript, and in the decision to publish the results.
Figures
References
-
- Vallianou N., Christodoulatos G.S., Karampela I., Tsilingiris D., Magkos F., Stratigou T., Kounatidis D., Dalamaga M. Understanding the role of the gut microbiome and microbial metabolites in non-alcoholic fatty liver disease: Current evidence and perspectives. Biomolecules. 2021;12:56. doi: 10.3390/biom12010056. - DOI - PMC - PubMed
-
- Kostic A.D., Gevers D., Siljander H., Vatanen T., Hyotylainen T., Hamalainen A.M., Peet A., Tillmann V., Poho P., Mattila I., et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

