Exploring the Solubility Limits of Edaravone in Neat Solvents and Binary Mixtures: Experimental and Machine Learning Study
- PMID: 37836720
- PMCID: PMC10574143
- DOI: 10.3390/molecules28196877
Exploring the Solubility Limits of Edaravone in Neat Solvents and Binary Mixtures: Experimental and Machine Learning Study
Abstract
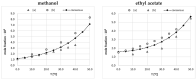
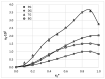
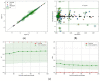
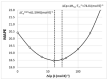
This study explores the edaravone solubility space encompassing both neat and binary dissolution media. Efforts were made to reveal the inherent concentration limits of common pure and mixed solvents. For this purpose, the published solubility data of the title drug were scrupulously inspected and cured, which made the dataset consistent and coherent. However, the lack of some important types of solvents in the collection called for an extension of the available pool of edaravone solubility data. Hence, new measurements were performed to collect edaravone solubility values in polar non-protic and diprotic media. Such an extended set of data was used in the machine learning process for tuning the parameters of regressor models and formulating the ensemble for predicting new data. In both phases, namely the model training and ensemble formulation, close attention was paid not only to minimizing the deviation of computed values from the experimental ones but also to ensuring high predictive power and accurate solubility computations for new systems. Furthermore, the environmental friendliness characteristics determined based on the common green solvent selection criteria, were included in the analysis. Our applied protocol led to the conclusion that the solubility space defined by ordinary solvents is limited, and it is unlikely to find solvents that are better suited for edaravone dissolution than those described in this manuscript. The theoretical framework presented in this study provides a precise guideline for conducting experiments, as well as saving time and resources in the pursuit of new findings.
Keywords: COSMO-RS; deep learning; edaravone; green solvents; hyperparameter tuning; learning curve analysis; solubility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Drugbank Edaravone. [(accessed on 28 November 2022)]. Available online: https://go.drugbank.com/drugs/DB12243.
LinkOut - more resources
Full Text Sources

