Protein retention in the endoplasmic reticulum rescues Aβ toxicity in Drosophila
- PMID: 37837732
- PMCID: PMC10940166
- DOI: 10.1016/j.neurobiolaging.2023.09.008
Protein retention in the endoplasmic reticulum rescues Aβ toxicity in Drosophila
Abstract
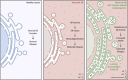
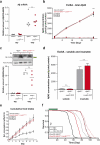
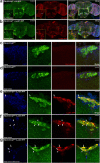
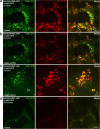
Amyloid β (Aβ) accumulation is a hallmark of Alzheimer's disease. In adult Drosophila brains, human Aβ overexpression harms climbing and lifespan. It's uncertain whether Aβ is intrinsically toxic or activates downstream neurodegeneration pathways. Our study uncovers a novel protective role against Aβ toxicity: intra-endoplasmic reticulum (ER) protein accumulation with a focus on laminin and collagen subunits. Despite high Aβ, laminin B1 (LanB1) overexpression robustly counters toxicity, suggesting a potential Aβ resistance mechanism. Other laminin subunits and collagen IV also alleviate Aβ toxicity; combining them with LanB1 augments the effect. Imaging reveals ER retention of LanB1 without altering Aβ secretion. LanB1's rescue function operates independently of the IRE1α/XBP1 ER stress response. ER-targeted GFP overexpression also mitigates Aβ toxicity, highlighting broader ER protein retention advantages. Proof-of-principle tests in murine hippocampal slices using mouse Lamb1 demonstrate ER retention in transduced cells, indicating a conserved mechanism. Though ER protein retention generally harms, it could paradoxically counter neuronal Aβ toxicity, offering a new therapeutic avenue for Alzheimer's disease.
Keywords: Alzheimer’s disease; Aβ toxicity; Drosophila melanogaster; ER retention; Endoplasmic reticulum; Laminin.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

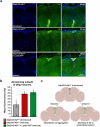
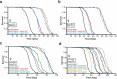
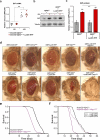
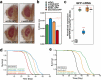

References
-
- Abramowski D., Rabe S., Upadhaya A.R., Reichwald J., Danner S., Staab D., Capetillo-Zarate E., Yamaguchi H., Saido T.C., Wiederhold K.-H., Thal D.R., Staufenbiel M. Transgenic expression of intraneuronal Aβ42 but not Aβ40 leads to cellular Aβ lesions, degeneration, and functional impairment without typical Alzheimer’s disease pathology. J. Neurosci. 2012;32:1273–1283. doi: 10.1523/jneurosci.4586-11.2012. - DOI - PMC - PubMed
-
- Arboleda-Velasquez J.F., Lopera F., O’Hare M., Delgado-Tirado S., Marino C., Chmielewska N., Saez-Torres K.L., Amarnani D., Schultz A.P., Sperling R.A., Leyton-Cifuentes D., Chen K., Baena A., Aguillon D., Rios-Romenets S., Giraldo M., Guzmán-Vélez E., Norton D.J., Pardilla-Delgado E., Artola A., Sanchez J.S., Acosta-Uribe J., Lalli M., Kosik K.S., Huentelman M.J., Zetterberg H., Blennow K., Reiman R.A., Luo J., Chen Y., Thiyyagura P., Su Y., Jun G.R., Naymik M., Gai X., Bootwalla M., Ji J., Shen L., Miller J.B., Kim L.A., Tariot P.N., Johnson K.A., Reiman E.M., Quiroz Y.T. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat. Med. 2019;25:1680–1683. doi: 10.1038/s41591-019-0611-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

