Cryptic susceptibility to penicillin/β-lactamase inhibitor combinations in emerging multidrug-resistant, hospital-adapted Staphylococcus epidermidis lineages
- PMID: 37838722
- PMCID: PMC10576800
- DOI: 10.1038/s41467-023-42245-y
Cryptic susceptibility to penicillin/β-lactamase inhibitor combinations in emerging multidrug-resistant, hospital-adapted Staphylococcus epidermidis lineages
Abstract
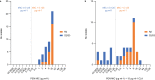
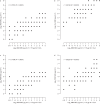
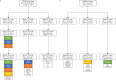
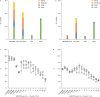
Global spread of multidrug-resistant, hospital-adapted Staphylococcus epidermidis lineages underscores the need for new therapeutic strategies. Here we show that many S. epidermidis isolates belonging to these lineages display cryptic susceptibility to penicillin/β-lactamase inhibitor combinations under in vitro conditions, despite carrying the methicillin resistance gene mecA. Using a mouse thigh model of S. epidermidis infection, we demonstrate that single-dose treatment with amoxicillin/clavulanic acid significantly reduces methicillin-resistant S. epidermidis loads without leading to detectable resistance development. On the other hand, we also show that methicillin-resistant S. epidermidis is capable of developing increased resistance to amoxicillin/clavulanic acid during long-term in vitro exposure to these drugs. These findings suggest that penicillin/β-lactamase inhibitor combinations could be a promising therapeutic candidate for treatment of a high proportion of methicillin-resistant S. epidermidis infections, although the in vivo risk of resistance development needs to be further addressed before they can be incorporated into clinical trials.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- European Centre for Disease Prevention and Control. Multidrug-resistant Staphylococcus epidermidis. https://www.ecdc.europa.eu/sites/default/files/documents/15-10-2018-RRA-... (2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

