Androgen receptor is a determinant of melanoma targeted drug resistance
- PMID: 37838724
- PMCID: PMC10576812
- DOI: 10.1038/s41467-023-42239-w
Androgen receptor is a determinant of melanoma targeted drug resistance
Abstract
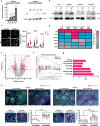
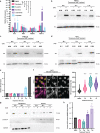
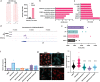
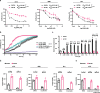
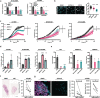
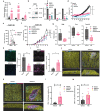
Melanoma provides a primary benchmark for targeted drug therapy. Most melanomas with BRAFV600 mutations regress in response to BRAF/MEK inhibitors (BRAFi/MEKi). However, nearly all relapse within the first two years, and there is a connection between BRAFi/MEKi-resistance and poor response to immune checkpoint therapy. We reported that androgen receptor (AR) activity is required for melanoma cell proliferation and tumorigenesis. We show here that AR expression is markedly increased in BRAFi-resistant melanoma cells, and in sensitive cells soon after BRAFi exposure. Increased AR expression is sufficient to render melanoma cells BRAFi-resistant, eliciting transcriptional changes of BRAFi-resistant subpopulations, including elevated EGFR and SERPINE1 expression, of likely clinical significance. Inhibition of AR expression or activity blunts changes in gene expression and suppresses proliferation and tumorigenesis of BRAFi-resistant melanoma cells, promoting clusters of CD8+ T cells infiltration and cancer cells killing. Our findings point to targeting AR as possible co-therapeutical approach in melanoma treatment.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gamba CS, Clarke CA, Keegan THM, Tao L, Swetter SM. Melanoma survival disadvantage in young, non-hispanic white males compared with females. Jama Dermatol. 2013;149:912–920. - PubMed
-
- Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nat. Rev. Cancer. 2016;16:330–339. - PubMed
-
- Nosrati A, Wei ML. Sex disparities in melanoma outcomes: the role of biology. Arch. Biochem Biophys. 2014;563:42–50. - PubMed
-
- Bartz D, et al. Clinical advances in sex- and gender-informed medicine to improve the health of all: a review. JAMA Intern. Med. 2020;180:574–583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

