MANF stimulates autophagy and restores mitochondrial homeostasis to treat autosomal dominant tubulointerstitial kidney disease in mice
- PMID: 37838725
- PMCID: PMC10576802
- DOI: 10.1038/s41467-023-42154-0
MANF stimulates autophagy and restores mitochondrial homeostasis to treat autosomal dominant tubulointerstitial kidney disease in mice
Abstract
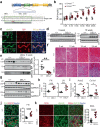
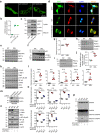
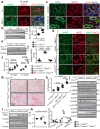
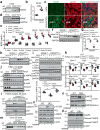
Misfolded protein aggregates may cause toxic proteinopathy, including autosomal dominant tubulointerstitial kidney disease due to uromodulin mutations (ADTKD-UMOD), a leading hereditary kidney disease. There are no targeted therapies. In our generated mouse model recapitulating human ADTKD-UMOD carrying a leading UMOD mutation, we show that autophagy/mitophagy and mitochondrial biogenesis are impaired, leading to cGAS-STING activation and tubular injury. Moreover, we demonstrate that inducible tubular overexpression of mesencephalic astrocyte-derived neurotrophic factor (MANF), a secreted endoplasmic reticulum protein, after the onset of disease stimulates autophagy/mitophagy, clears mutant UMOD, and promotes mitochondrial biogenesis through p-AMPK enhancement, thus protecting kidney function in our ADTKD mouse model. Conversely, genetic ablation of MANF in the mutant thick ascending limb tubular cells worsens autophagy suppression and kidney fibrosis. Together, we have discovered MANF as a biotherapeutic protein and elucidated previously unknown mechanisms of MANF in the regulation of organelle homeostasis, which may have broad therapeutic applications to treat various proteinopathies.
© 2023. Springer Nature Limited.
Conflict of interest statement
Y.M.C., S.J.P., Y.K., and F.U. are inventors on a patent entitled “Compositions and methods for treating and preventing endoplasmic reticulum (ER) stress-mediated kidney diseases” (US 11,129,871), which was issued by US Patent and Trademark Office in Sep. 2021. Y.M.C. and Y.K. are inventors on a patent entitled “methods of detecting biomarkers of endoplasmic reticulum (ER) stress-associated kidney diseases” (US 10,156,564), which was issued by US Patent and Trademark Office on Dec. 18, 2018. J.S. and V.S. are inventors on a non-provisional patent, entitled “PET tracers for noninvasive imaging of ROS activity” filed by Washington University in St. Louis, St. Louis, MO. Authors (J.S. & V.S.) declare no competing interests. The remaining authors declare no competing interests.
Figures




Update of
-
MANF stimulates autophagy and restores mitochondrial homeostasis to treat toxic proteinopathy.bioRxiv [Preprint]. 2023 Jan 12:2023.01.10.523171. doi: 10.1101/2023.01.10.523171. bioRxiv. 2023. Update in: Nat Commun. 2023 Oct 14;14(1):6493. doi: 10.1038/s41467-023-42154-0. PMID: 36711449 Free PMC article. Updated. Preprint.
References
-
- Walker AK, Atkin JD. Stress signaling from the endoplasmic reticulum: a central player in the pathogenesis of amyotrophic lateral sclerosis. IUBMB Life. 2011;63:754–763. - PubMed
-
- Howard M, Welch WJ. Manipulating the folding pathway of delta F508 CFTR using chemical chaperones. Methods Mol. Med. 2002;70:267–275. - PubMed
-
- Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY025799/EY/NEI NIH HHS/United States
- R01 HL111163/HL/NHLBI NIH HHS/United States
- U54 DK137307/DK/NIDDK NIH HHS/United States
- P41 EB025815/EB/NIBIB NIH HHS/United States
- R35 GM128772/GM/NIGMS NIH HHS/United States
- R01 DK132090/DK/NIDDK NIH HHS/United States
- R21 DK106584/DK/NIDDK NIH HHS/United States
- R03 DK106451/DK/NIDDK NIH HHS/United States
- R21 DK131557/DK/NIDDK NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- R01 DK105056/DK/NIDDK NIH HHS/United States
- P30 DK020579/DK/NIDDK NIH HHS/United States
- P30 DK114857/DK/NIDDK NIH HHS/United States
- K08 DK089015/DK/NIDDK NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- R01 HL142297/HL/NHLBI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- P30 DK079337/DK/NIDDK NIH HHS/United States
- P30 AR057235/AR/NIAMS NIH HHS/United States
- R01 DK112921/DK/NIDDK NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- UH2 TR002065/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

