Domain architecture and protein-protein interactions regulate KDM5A recruitment to the chromatin
- PMID: 37838974
- PMCID: PMC10578193
- DOI: 10.1080/15592294.2023.2268813
Domain architecture and protein-protein interactions regulate KDM5A recruitment to the chromatin
Abstract
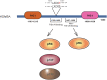
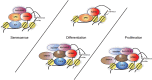
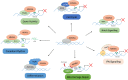
Tri-methylation of Histone 3 lysine 4 (H3K4) is an important epigenetic modification whose deposition and removal can affect the chromatin at structural and functional levels. KDM5A is one of the four known H3K4-specific demethylases. It is a part of the KDM5 family, which is characterized by a catalytic Jumonji domain capable of removing H3K4 di- and tri-methylation marks. KDM5A has been found to be involved in multiple cellular processes such as differentiation, metabolism, cell cycle, and transcription. Its link to various diseases, including cancer, makes KDM5A an important target for drug development. However, despite several studies outlining its significance in various pathways, our lack of understanding of its recruitment and function at the target sites on the chromatin presents a challenge in creating effective and targeted treatments. Therefore, it is essential to understand the recruitment mechanism of KDM5A to chromatin, and its activity therein, to comprehend how various roles of KDM5A are regulated. In this review, we discuss how KDM5A functions in a context-dependent manner on the chromatin, either directly through its structural domain, or through various interacting partners, to bring about a diverse range of functions.
Keywords: ARID domain; KDM5A; Plant Homeodomain; chromatin recruitment; demethylase; pocket proteins.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
