Epigenetic regulation of TP53 is involved in prostate cancer radioresistance and DNA damage response signaling
- PMID: 37840069
- PMCID: PMC10577134
- DOI: 10.1038/s41392-023-01639-6
Epigenetic regulation of TP53 is involved in prostate cancer radioresistance and DNA damage response signaling
Abstract
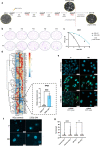
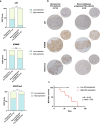
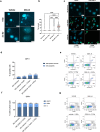
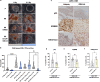
External beam radiotherapy (RT) is a leading first-line therapy for prostate cancer (PCa), and, in recent years, significant advances have been accomplished. However, RT resistance can arise and result in long-term recurrence or disease progression in the worst-case scenario. Thus, making crucial the discovery of new targets for PCa radiosensitization. Herein, we generated a radioresistant PCa cell line, and found p53 to be highly expressed in radioresistant PCa cells, as well as in PCa patients with recurrent/disease progression submitted to RT. Mechanism dissection revealed that RT could promote p53 expression via epigenetic modulation. Specifically, a decrease of H3K27me3 occupancy at TP53 gene promoter, due to increased KDM6B activity, was observed in radioresistant PCa cells. Furthermore, p53 is essential for efficient DNA damage signaling response and cell recovery upon stress induction by prolonged fractionated irradiation. Remarkably, KDM6B inhibition by GSK-J4 significantly decreased p53 expression, consequently attenuating the radioresistant phenotype of PCa cells and hampering in vivo 3D tumor formation. Overall, this work contributes to improve the understanding of p53 as a mediator of signaling transduction in DNA damage repair, as well as the impact of epigenetic targeting for PCa radiosensitization.
© 2023. West China Hospital, Sichuan University.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

