Development of 12 sets of chromosome segment substitution lines that enhance allele mining in Asian cultivated rice
- PMID: 37840983
- PMCID: PMC10570878
- DOI: 10.1270/jsbbs.23006
Development of 12 sets of chromosome segment substitution lines that enhance allele mining in Asian cultivated rice
Abstract
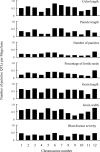
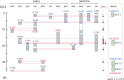
Many agronomic traits that are important in rice breeding are controlled by multiple genes. The extensive time and effort devoted so far to identifying and selecting such genes are still not enough to target multiple agronomic traits in practical breeding in Japan because of a lack of suitable plant materials in which to efficiently detect and validate beneficial alleles from diverse genetic resources. To facilitate the comprehensive analysis of genetic variation in agronomic traits among Asian cultivated rice, we developed 12 sets of chromosome segment substitution lines (CSSLs) with the japonica background, 11 of them in the same genetic background, using donors representing the genetic diversity of Asian cultivated rice. Using these materials, we overviewed the chromosomal locations of 1079 putative QTLs for seven agronomic traits and their allelic distribution in Asian cultivated rice through multiple linear regression analysis. The CSSLs will allow the effects of putative QTLs in the highly homogeneous japonica background to be validated.
Keywords: CSSLs; Oryza sativa L.; advanced mapping population; allele mining.
Copyright © 2023 by JAPANESE SOCIETY OF BREEDING.
Figures

References
-
- Ando, T., Yamamoto T., Shimizu T., Ma X.F., Shomura A., Takeuchi Y., Lin S.Y. and Yano M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor Appl Genet 116: 881–890. - PubMed
-
- Arbelaez, J.D., Moreno L.T., Singh N., Tung C.W., Maron L.G., Ospina Y., Martinez C.P., Grenier C., Lorieux M. and McCouch S. (2015) Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent, O. sativa cv. Curinga. Mol Breed 35: 81. - PMC - PubMed
-
- Asaga, K. (1976) Evaluation standard of rice leaf blast in nursery test. J Agric Sci 31: 156–156 (in Japanese).
-
- Balakrishnan, D., Surapaneni M., Mesapogu S. and Neelamraju S. (2019) Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor Appl Genet 132: 1–25. - PubMed

